
Pelargonium line pattern virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Pelarspovirus
Average proteome isoelectric point is 8.29
Get precalculated fractions of proteins
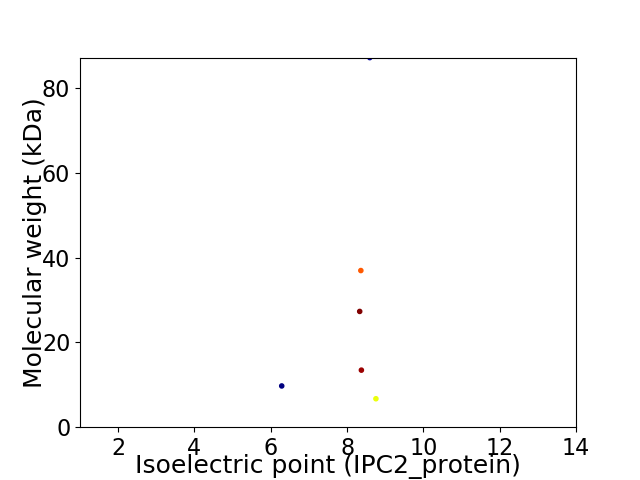
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5XKN5|U5XKN5_9TOMB p9.7 OS=Pelargonium line pattern virus OX=167019 GN=p9.7 PE=4 SV=1
MM1 pKa = 7.89EE2 pKa = 4.57YY3 pKa = 10.27PRR5 pKa = 11.84VHH7 pKa = 6.62LAILSVLISSQLLIKK22 pKa = 9.6WNLWSISISDD32 pKa = 4.8FLPQPHH38 pKa = 6.55SLHH41 pKa = 6.85PNLLVCIVLCIFFSSVLSQGQSYY64 pKa = 10.14SYY66 pKa = 11.21SYY68 pKa = 11.15FSTSTSDD75 pKa = 3.02KK76 pKa = 10.0FISVAVGNGGQGG88 pKa = 2.97
MM1 pKa = 7.89EE2 pKa = 4.57YY3 pKa = 10.27PRR5 pKa = 11.84VHH7 pKa = 6.62LAILSVLISSQLLIKK22 pKa = 9.6WNLWSISISDD32 pKa = 4.8FLPQPHH38 pKa = 6.55SLHH41 pKa = 6.85PNLLVCIVLCIFFSSVLSQGQSYY64 pKa = 10.14SYY66 pKa = 11.21SYY68 pKa = 11.15FSTSTSDD75 pKa = 3.02KK76 pKa = 10.0FISVAVGNGGQGG88 pKa = 2.97
Molecular weight: 9.74 kDa
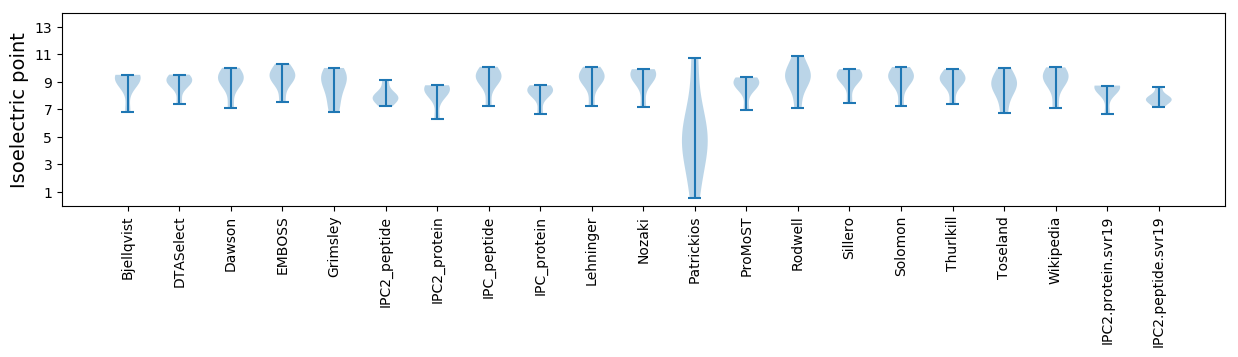
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5XKN5|U5XKN5_9TOMB p9.7 OS=Pelargonium line pattern virus OX=167019 GN=p9.7 PE=4 SV=1
MM1 pKa = 7.24AHH3 pKa = 6.24YY4 pKa = 10.09FGEE7 pKa = 4.27ALQLGLFVAKK17 pKa = 10.43ASCWASKK24 pKa = 10.32EE25 pKa = 3.91LALLGPNLAWIATRR39 pKa = 11.84PLRR42 pKa = 11.84NDD44 pKa = 2.62ANAILKK50 pKa = 10.38RR51 pKa = 11.84IGVHH55 pKa = 6.09GLAKK59 pKa = 10.11IPCNFDD65 pKa = 3.31SDD67 pKa = 4.21EE68 pKa = 4.01LTIANDD74 pKa = 4.52CIVKK78 pKa = 10.05SDD80 pKa = 3.91RR81 pKa = 11.84NLEE84 pKa = 4.16DD85 pKa = 3.23EE86 pKa = 4.77SIVVEE91 pKa = 4.23EE92 pKa = 4.31VEE94 pKa = 4.27EE95 pKa = 4.74GEE97 pKa = 4.29EE98 pKa = 4.02KK99 pKa = 10.62KK100 pKa = 10.55KK101 pKa = 10.75SKK103 pKa = 10.39KK104 pKa = 5.49VTRR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84VKK111 pKa = 9.8TKK113 pKa = 8.61PTFAAVLAADD123 pKa = 3.97AKK125 pKa = 10.77NYY127 pKa = 9.63YY128 pKa = 10.16GCLPSATRR136 pKa = 11.84ANEE139 pKa = 3.92LSVMKK144 pKa = 10.33YY145 pKa = 10.09LVSKK149 pKa = 10.14CQEE152 pKa = 3.84HH153 pKa = 7.16KK154 pKa = 10.55LTITQTRR161 pKa = 11.84EE162 pKa = 3.4VSAMAFALTFTPDD175 pKa = 3.24EE176 pKa = 4.14NDD178 pKa = 3.19KK179 pKa = 11.35LIYY182 pKa = 10.08KK183 pKa = 8.8YY184 pKa = 11.06LNSTEE189 pKa = 4.09VFEE192 pKa = 5.68RR193 pKa = 11.84RR194 pKa = 11.84VDD196 pKa = 3.49YY197 pKa = 11.25AKK199 pKa = 10.97ARR201 pKa = 11.84GVDD204 pKa = 3.48KK205 pKa = 11.01CWFEE209 pKa = 3.79LLKK212 pKa = 10.83RR213 pKa = 11.84PWHH216 pKa = 5.1ARR218 pKa = 11.84AWRR221 pKa = 11.84RR222 pKa = 11.84VVGRR226 pKa = 11.84IFGLPEE232 pKa = 3.33QQAFEE237 pKa = 4.2FVKK240 pKa = 9.48XGCLMDD246 pKa = 3.51TCGVDD251 pKa = 3.08TKK253 pKa = 10.75VYY255 pKa = 9.7RR256 pKa = 11.84GEE258 pKa = 4.04HH259 pKa = 4.88RR260 pKa = 11.84WVKK263 pKa = 9.32EE264 pKa = 3.48FRR266 pKa = 11.84GAANPKK272 pKa = 9.12PRR274 pKa = 11.84RR275 pKa = 11.84LYY277 pKa = 10.3KK278 pKa = 10.25ISGVSPEE285 pKa = 4.3VRR287 pKa = 11.84WGVHH291 pKa = 4.89NNSFVNLRR299 pKa = 11.84RR300 pKa = 11.84GLMEE304 pKa = 3.41RR305 pKa = 11.84VFYY308 pKa = 10.13VEE310 pKa = 4.83RR311 pKa = 11.84SGEE314 pKa = 4.12LLPCPSPEE322 pKa = 3.79AGLFKK327 pKa = 10.86RR328 pKa = 11.84LFNKK332 pKa = 9.53VGRR335 pKa = 11.84RR336 pKa = 11.84VIRR339 pKa = 11.84FCGHH343 pKa = 6.88HH344 pKa = 6.24SPIPRR349 pKa = 11.84ASYY352 pKa = 9.84PGMFQGRR359 pKa = 11.84KK360 pKa = 5.56RR361 pKa = 11.84TIYY364 pKa = 9.64EE365 pKa = 3.67NAVRR369 pKa = 11.84SLVDD373 pKa = 3.11RR374 pKa = 11.84PYY376 pKa = 11.01NIRR379 pKa = 11.84DD380 pKa = 3.49SYY382 pKa = 11.74LKK384 pKa = 10.07TFVKK388 pKa = 10.58FEE390 pKa = 4.24KK391 pKa = 10.63LDD393 pKa = 3.82FSKK396 pKa = 11.03KK397 pKa = 9.08PDD399 pKa = 3.82PAPRR403 pKa = 11.84VIQPRR408 pKa = 11.84HH409 pKa = 4.77PRR411 pKa = 11.84YY412 pKa = 9.36NVEE415 pKa = 3.88LGRR418 pKa = 11.84YY419 pKa = 7.19LKK421 pKa = 10.46PFEE424 pKa = 4.32HH425 pKa = 6.76FCYY428 pKa = 10.23KK429 pKa = 10.7ALDD432 pKa = 4.05KK433 pKa = 11.12LWGGPTVMKK442 pKa = 10.44GYY444 pKa = 8.15TVEE447 pKa = 4.02EE448 pKa = 3.8MGGIIKK454 pKa = 10.23DD455 pKa = 3.12SWLQFQKK462 pKa = 10.59PVAIGFDD469 pKa = 3.3MSRR472 pKa = 11.84FDD474 pKa = 3.24QHH476 pKa = 8.4VSVDD480 pKa = 3.42ALKK483 pKa = 10.67FEE485 pKa = 4.16HH486 pKa = 6.96SIYY489 pKa = 10.24KK490 pKa = 10.43ACFSKK495 pKa = 10.89DD496 pKa = 3.17GNLATLLGHH505 pKa = 6.42QISNRR510 pKa = 11.84GSAYY514 pKa = 10.57ANDD517 pKa = 4.32GYY519 pKa = 11.23LRR521 pKa = 11.84YY522 pKa = 9.38RR523 pKa = 11.84VEE525 pKa = 4.38GKK527 pKa = 10.57RR528 pKa = 11.84MSGDD532 pKa = 3.26VNTGLGNCLLACTITKK548 pKa = 10.2FLMEE552 pKa = 4.89EE553 pKa = 3.38IGVRR557 pKa = 11.84SRR559 pKa = 11.84LVNNGDD565 pKa = 3.76DD566 pKa = 3.67CVLICEE572 pKa = 4.96AGDD575 pKa = 3.59CRR577 pKa = 11.84AVEE580 pKa = 4.35GSLTVGWRR588 pKa = 11.84RR589 pKa = 11.84FGFTCIAEE597 pKa = 4.24KK598 pKa = 10.15PVYY601 pKa = 9.24EE602 pKa = 4.02LEE604 pKa = 4.78KK605 pKa = 10.64IEE607 pKa = 4.29FCQMSPIQISDD618 pKa = 3.43SRR620 pKa = 11.84VKK622 pKa = 10.53LVRR625 pKa = 11.84KK626 pKa = 7.02PQKK629 pKa = 10.54SISKK633 pKa = 8.91DD634 pKa = 3.23AHH636 pKa = 5.36STTPLTTIQLAQEE649 pKa = 3.78WTRR652 pKa = 11.84AIGEE656 pKa = 4.5GGLSLTSGIPVVQEE670 pKa = 4.63FYY672 pKa = 11.01QCLIRR677 pKa = 11.84NGKK680 pKa = 9.1KK681 pKa = 8.84ATKK684 pKa = 9.04EE685 pKa = 4.05SKK687 pKa = 10.74KK688 pKa = 9.02MAFYY692 pKa = 11.13GDD694 pKa = 5.06YY695 pKa = 8.73YY696 pKa = 9.57WKK698 pKa = 9.91WVNQHH703 pKa = 5.2SGKK706 pKa = 10.34YY707 pKa = 9.5EE708 pKa = 3.83PVTEE712 pKa = 4.17EE713 pKa = 3.93ARR715 pKa = 11.84HH716 pKa = 5.4SFHH719 pKa = 7.01LAFGVSPDD727 pKa = 3.49QQLALEE733 pKa = 5.16DD734 pKa = 3.38IYY736 pKa = 11.6SRR738 pKa = 11.84KK739 pKa = 9.61SLLWEE744 pKa = 3.93SSPFTFEE751 pKa = 3.31VGAIEE756 pKa = 4.55WIFNQKK762 pKa = 9.56IPII765 pKa = 3.98
MM1 pKa = 7.24AHH3 pKa = 6.24YY4 pKa = 10.09FGEE7 pKa = 4.27ALQLGLFVAKK17 pKa = 10.43ASCWASKK24 pKa = 10.32EE25 pKa = 3.91LALLGPNLAWIATRR39 pKa = 11.84PLRR42 pKa = 11.84NDD44 pKa = 2.62ANAILKK50 pKa = 10.38RR51 pKa = 11.84IGVHH55 pKa = 6.09GLAKK59 pKa = 10.11IPCNFDD65 pKa = 3.31SDD67 pKa = 4.21EE68 pKa = 4.01LTIANDD74 pKa = 4.52CIVKK78 pKa = 10.05SDD80 pKa = 3.91RR81 pKa = 11.84NLEE84 pKa = 4.16DD85 pKa = 3.23EE86 pKa = 4.77SIVVEE91 pKa = 4.23EE92 pKa = 4.31VEE94 pKa = 4.27EE95 pKa = 4.74GEE97 pKa = 4.29EE98 pKa = 4.02KK99 pKa = 10.62KK100 pKa = 10.55KK101 pKa = 10.75SKK103 pKa = 10.39KK104 pKa = 5.49VTRR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84VKK111 pKa = 9.8TKK113 pKa = 8.61PTFAAVLAADD123 pKa = 3.97AKK125 pKa = 10.77NYY127 pKa = 9.63YY128 pKa = 10.16GCLPSATRR136 pKa = 11.84ANEE139 pKa = 3.92LSVMKK144 pKa = 10.33YY145 pKa = 10.09LVSKK149 pKa = 10.14CQEE152 pKa = 3.84HH153 pKa = 7.16KK154 pKa = 10.55LTITQTRR161 pKa = 11.84EE162 pKa = 3.4VSAMAFALTFTPDD175 pKa = 3.24EE176 pKa = 4.14NDD178 pKa = 3.19KK179 pKa = 11.35LIYY182 pKa = 10.08KK183 pKa = 8.8YY184 pKa = 11.06LNSTEE189 pKa = 4.09VFEE192 pKa = 5.68RR193 pKa = 11.84RR194 pKa = 11.84VDD196 pKa = 3.49YY197 pKa = 11.25AKK199 pKa = 10.97ARR201 pKa = 11.84GVDD204 pKa = 3.48KK205 pKa = 11.01CWFEE209 pKa = 3.79LLKK212 pKa = 10.83RR213 pKa = 11.84PWHH216 pKa = 5.1ARR218 pKa = 11.84AWRR221 pKa = 11.84RR222 pKa = 11.84VVGRR226 pKa = 11.84IFGLPEE232 pKa = 3.33QQAFEE237 pKa = 4.2FVKK240 pKa = 9.48XGCLMDD246 pKa = 3.51TCGVDD251 pKa = 3.08TKK253 pKa = 10.75VYY255 pKa = 9.7RR256 pKa = 11.84GEE258 pKa = 4.04HH259 pKa = 4.88RR260 pKa = 11.84WVKK263 pKa = 9.32EE264 pKa = 3.48FRR266 pKa = 11.84GAANPKK272 pKa = 9.12PRR274 pKa = 11.84RR275 pKa = 11.84LYY277 pKa = 10.3KK278 pKa = 10.25ISGVSPEE285 pKa = 4.3VRR287 pKa = 11.84WGVHH291 pKa = 4.89NNSFVNLRR299 pKa = 11.84RR300 pKa = 11.84GLMEE304 pKa = 3.41RR305 pKa = 11.84VFYY308 pKa = 10.13VEE310 pKa = 4.83RR311 pKa = 11.84SGEE314 pKa = 4.12LLPCPSPEE322 pKa = 3.79AGLFKK327 pKa = 10.86RR328 pKa = 11.84LFNKK332 pKa = 9.53VGRR335 pKa = 11.84RR336 pKa = 11.84VIRR339 pKa = 11.84FCGHH343 pKa = 6.88HH344 pKa = 6.24SPIPRR349 pKa = 11.84ASYY352 pKa = 9.84PGMFQGRR359 pKa = 11.84KK360 pKa = 5.56RR361 pKa = 11.84TIYY364 pKa = 9.64EE365 pKa = 3.67NAVRR369 pKa = 11.84SLVDD373 pKa = 3.11RR374 pKa = 11.84PYY376 pKa = 11.01NIRR379 pKa = 11.84DD380 pKa = 3.49SYY382 pKa = 11.74LKK384 pKa = 10.07TFVKK388 pKa = 10.58FEE390 pKa = 4.24KK391 pKa = 10.63LDD393 pKa = 3.82FSKK396 pKa = 11.03KK397 pKa = 9.08PDD399 pKa = 3.82PAPRR403 pKa = 11.84VIQPRR408 pKa = 11.84HH409 pKa = 4.77PRR411 pKa = 11.84YY412 pKa = 9.36NVEE415 pKa = 3.88LGRR418 pKa = 11.84YY419 pKa = 7.19LKK421 pKa = 10.46PFEE424 pKa = 4.32HH425 pKa = 6.76FCYY428 pKa = 10.23KK429 pKa = 10.7ALDD432 pKa = 4.05KK433 pKa = 11.12LWGGPTVMKK442 pKa = 10.44GYY444 pKa = 8.15TVEE447 pKa = 4.02EE448 pKa = 3.8MGGIIKK454 pKa = 10.23DD455 pKa = 3.12SWLQFQKK462 pKa = 10.59PVAIGFDD469 pKa = 3.3MSRR472 pKa = 11.84FDD474 pKa = 3.24QHH476 pKa = 8.4VSVDD480 pKa = 3.42ALKK483 pKa = 10.67FEE485 pKa = 4.16HH486 pKa = 6.96SIYY489 pKa = 10.24KK490 pKa = 10.43ACFSKK495 pKa = 10.89DD496 pKa = 3.17GNLATLLGHH505 pKa = 6.42QISNRR510 pKa = 11.84GSAYY514 pKa = 10.57ANDD517 pKa = 4.32GYY519 pKa = 11.23LRR521 pKa = 11.84YY522 pKa = 9.38RR523 pKa = 11.84VEE525 pKa = 4.38GKK527 pKa = 10.57RR528 pKa = 11.84MSGDD532 pKa = 3.26VNTGLGNCLLACTITKK548 pKa = 10.2FLMEE552 pKa = 4.89EE553 pKa = 3.38IGVRR557 pKa = 11.84SRR559 pKa = 11.84LVNNGDD565 pKa = 3.76DD566 pKa = 3.67CVLICEE572 pKa = 4.96AGDD575 pKa = 3.59CRR577 pKa = 11.84AVEE580 pKa = 4.35GSLTVGWRR588 pKa = 11.84RR589 pKa = 11.84FGFTCIAEE597 pKa = 4.24KK598 pKa = 10.15PVYY601 pKa = 9.24EE602 pKa = 4.02LEE604 pKa = 4.78KK605 pKa = 10.64IEE607 pKa = 4.29FCQMSPIQISDD618 pKa = 3.43SRR620 pKa = 11.84VKK622 pKa = 10.53LVRR625 pKa = 11.84KK626 pKa = 7.02PQKK629 pKa = 10.54SISKK633 pKa = 8.91DD634 pKa = 3.23AHH636 pKa = 5.36STTPLTTIQLAQEE649 pKa = 3.78WTRR652 pKa = 11.84AIGEE656 pKa = 4.5GGLSLTSGIPVVQEE670 pKa = 4.63FYY672 pKa = 11.01QCLIRR677 pKa = 11.84NGKK680 pKa = 9.1KK681 pKa = 8.84ATKK684 pKa = 9.04EE685 pKa = 4.05SKK687 pKa = 10.74KK688 pKa = 9.02MAFYY692 pKa = 11.13GDD694 pKa = 5.06YY695 pKa = 8.73YY696 pKa = 9.57WKK698 pKa = 9.91WVNQHH703 pKa = 5.2SGKK706 pKa = 10.34YY707 pKa = 9.5EE708 pKa = 3.83PVTEE712 pKa = 4.17EE713 pKa = 3.93ARR715 pKa = 11.84HH716 pKa = 5.4SFHH719 pKa = 7.01LAFGVSPDD727 pKa = 3.49QQLALEE733 pKa = 5.16DD734 pKa = 3.38IYY736 pKa = 11.6SRR738 pKa = 11.84KK739 pKa = 9.61SLLWEE744 pKa = 3.93SSPFTFEE751 pKa = 3.31VGAIEE756 pKa = 4.55WIFNQKK762 pKa = 9.56IPII765 pKa = 3.98
Molecular weight: 87.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1621 |
63 |
765 |
270.2 |
30.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.588 ± 0.984 | 2.468 ± 0.322 |
4.257 ± 0.286 | 6.416 ± 1.019 |
4.257 ± 0.627 | 6.971 ± 0.802 |
2.097 ± 0.2 | 4.812 ± 0.422 |
7.65 ± 0.868 | 8.945 ± 0.601 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.604 ± 0.197 | 3.825 ± 0.291 |
4.257 ± 0.51 | 2.714 ± 0.291 |
6.477 ± 0.915 | 8.143 ± 2.13 |
5.12 ± 0.655 | 7.341 ± 0.588 |
1.727 ± 0.346 | 3.27 ± 0.492 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
