
Avian paramyxovirus 15
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Metaavulavirus; Avian metaavulavirus 15
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
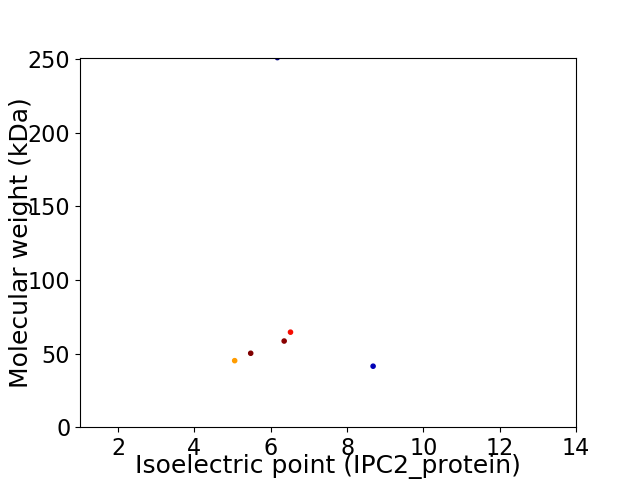
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6R4X6|A0A1W6R4X6_9MONO Nucleocapsid OS=Avian paramyxovirus 15 OX=1983777 GN=NP PE=3 SV=1
MM1 pKa = 7.48EE2 pKa = 5.32FKK4 pKa = 10.59SDD6 pKa = 3.85EE7 pKa = 4.6EE8 pKa = 4.66INDD11 pKa = 4.12LLNLSSSVISEE22 pKa = 4.02LQRR25 pKa = 11.84AEE27 pKa = 4.3LKK29 pKa = 10.66PPQTLGKK36 pKa = 8.76PQVPSGNTTSLISLWEE52 pKa = 4.06AEE54 pKa = 4.25GAPEE58 pKa = 4.14NKK60 pKa = 9.73LGRR63 pKa = 11.84GNTGTATDD71 pKa = 3.91TEE73 pKa = 4.82GEE75 pKa = 3.89QDD77 pKa = 3.49QQIVQTEE84 pKa = 4.12IEE86 pKa = 4.22QTHH89 pKa = 6.15TEE91 pKa = 4.11DD92 pKa = 3.48TCPKK96 pKa = 9.38EE97 pKa = 4.2IPVDD101 pKa = 3.68NPGSFTQEE109 pKa = 3.24SDD111 pKa = 3.74LDD113 pKa = 3.72KK114 pKa = 10.74TLKK117 pKa = 10.71KK118 pKa = 10.62LEE120 pKa = 4.02QRR122 pKa = 11.84NSNLLKK128 pKa = 10.15KK129 pKa = 9.03TDD131 pKa = 3.73SPDD134 pKa = 3.1AGTTFKK140 pKa = 10.84KK141 pKa = 10.52GGQIAPKK148 pKa = 9.37HH149 pKa = 5.49QPLPSQVIEE158 pKa = 4.51GNLPPQDD165 pKa = 3.94CRR167 pKa = 11.84QSRR170 pKa = 11.84PSRR173 pKa = 11.84TDD175 pKa = 2.66ASQRR179 pKa = 11.84PSTTPKK185 pKa = 9.69GAHH188 pKa = 6.18PQLHH192 pKa = 6.78PFQDD196 pKa = 3.59IEE198 pKa = 4.56EE199 pKa = 4.36NTQSVPLDD207 pKa = 3.74HH208 pKa = 7.16PLKK211 pKa = 10.31LLVGATPDD219 pKa = 3.18VHH221 pKa = 7.85QFEE224 pKa = 4.58QSQGEE229 pKa = 4.32NGAHH233 pKa = 5.65VGNVLEE239 pKa = 4.37SASFAEE245 pKa = 4.41MTLSVLNEE253 pKa = 3.51VLIRR257 pKa = 11.84VARR260 pKa = 11.84MEE262 pKa = 4.15EE263 pKa = 4.17KK264 pKa = 10.61INDD267 pKa = 3.55ILKK270 pKa = 9.19TNSTIPLIRR279 pKa = 11.84NDD281 pKa = 3.0ISQLKK286 pKa = 8.45ATTALLSTQMASVQVLDD303 pKa = 4.39PGNAGFKK310 pKa = 10.51SLSEE314 pKa = 3.99MKK316 pKa = 10.34AASKK320 pKa = 9.74PAIIAISGPGDD331 pKa = 3.46MDD333 pKa = 4.27AVPIQDD339 pKa = 3.67KK340 pKa = 11.14LLVKK344 pKa = 10.49DD345 pKa = 3.51VLGRR349 pKa = 11.84PISAEE354 pKa = 3.68RR355 pKa = 11.84NIQAKK360 pKa = 7.11EE361 pKa = 4.25TPDD364 pKa = 3.59SVITQSDD371 pKa = 3.06KK372 pKa = 11.57DD373 pKa = 4.29AIQSLIDD380 pKa = 3.59TLVEE384 pKa = 4.14DD385 pKa = 3.83TSKK388 pKa = 10.59QARR391 pKa = 11.84LKK393 pKa = 10.37AQLEE397 pKa = 4.3RR398 pKa = 11.84VQDD401 pKa = 3.66KK402 pKa = 10.98PGLLKK407 pKa = 10.66LKK409 pKa = 10.38RR410 pKa = 11.84LIYY413 pKa = 10.29NAA415 pKa = 4.23
MM1 pKa = 7.48EE2 pKa = 5.32FKK4 pKa = 10.59SDD6 pKa = 3.85EE7 pKa = 4.6EE8 pKa = 4.66INDD11 pKa = 4.12LLNLSSSVISEE22 pKa = 4.02LQRR25 pKa = 11.84AEE27 pKa = 4.3LKK29 pKa = 10.66PPQTLGKK36 pKa = 8.76PQVPSGNTTSLISLWEE52 pKa = 4.06AEE54 pKa = 4.25GAPEE58 pKa = 4.14NKK60 pKa = 9.73LGRR63 pKa = 11.84GNTGTATDD71 pKa = 3.91TEE73 pKa = 4.82GEE75 pKa = 3.89QDD77 pKa = 3.49QQIVQTEE84 pKa = 4.12IEE86 pKa = 4.22QTHH89 pKa = 6.15TEE91 pKa = 4.11DD92 pKa = 3.48TCPKK96 pKa = 9.38EE97 pKa = 4.2IPVDD101 pKa = 3.68NPGSFTQEE109 pKa = 3.24SDD111 pKa = 3.74LDD113 pKa = 3.72KK114 pKa = 10.74TLKK117 pKa = 10.71KK118 pKa = 10.62LEE120 pKa = 4.02QRR122 pKa = 11.84NSNLLKK128 pKa = 10.15KK129 pKa = 9.03TDD131 pKa = 3.73SPDD134 pKa = 3.1AGTTFKK140 pKa = 10.84KK141 pKa = 10.52GGQIAPKK148 pKa = 9.37HH149 pKa = 5.49QPLPSQVIEE158 pKa = 4.51GNLPPQDD165 pKa = 3.94CRR167 pKa = 11.84QSRR170 pKa = 11.84PSRR173 pKa = 11.84TDD175 pKa = 2.66ASQRR179 pKa = 11.84PSTTPKK185 pKa = 9.69GAHH188 pKa = 6.18PQLHH192 pKa = 6.78PFQDD196 pKa = 3.59IEE198 pKa = 4.56EE199 pKa = 4.36NTQSVPLDD207 pKa = 3.74HH208 pKa = 7.16PLKK211 pKa = 10.31LLVGATPDD219 pKa = 3.18VHH221 pKa = 7.85QFEE224 pKa = 4.58QSQGEE229 pKa = 4.32NGAHH233 pKa = 5.65VGNVLEE239 pKa = 4.37SASFAEE245 pKa = 4.41MTLSVLNEE253 pKa = 3.51VLIRR257 pKa = 11.84VARR260 pKa = 11.84MEE262 pKa = 4.15EE263 pKa = 4.17KK264 pKa = 10.61INDD267 pKa = 3.55ILKK270 pKa = 9.19TNSTIPLIRR279 pKa = 11.84NDD281 pKa = 3.0ISQLKK286 pKa = 8.45ATTALLSTQMASVQVLDD303 pKa = 4.39PGNAGFKK310 pKa = 10.51SLSEE314 pKa = 3.99MKK316 pKa = 10.34AASKK320 pKa = 9.74PAIIAISGPGDD331 pKa = 3.46MDD333 pKa = 4.27AVPIQDD339 pKa = 3.67KK340 pKa = 11.14LLVKK344 pKa = 10.49DD345 pKa = 3.51VLGRR349 pKa = 11.84PISAEE354 pKa = 3.68RR355 pKa = 11.84NIQAKK360 pKa = 7.11EE361 pKa = 4.25TPDD364 pKa = 3.59SVITQSDD371 pKa = 3.06KK372 pKa = 11.57DD373 pKa = 4.29AIQSLIDD380 pKa = 3.59TLVEE384 pKa = 4.14DD385 pKa = 3.83TSKK388 pKa = 10.59QARR391 pKa = 11.84LKK393 pKa = 10.37AQLEE397 pKa = 4.3RR398 pKa = 11.84VQDD401 pKa = 3.66KK402 pKa = 10.98PGLLKK407 pKa = 10.66LKK409 pKa = 10.38RR410 pKa = 11.84LIYY413 pKa = 10.29NAA415 pKa = 4.23
Molecular weight: 45.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6R4T0|A0A1W6R4T0_9MONO Fusion glycoprotein F0 OS=Avian paramyxovirus 15 OX=1983777 GN=F PE=3 SV=1
MM1 pKa = 7.85ASTSIPLHH9 pKa = 6.37INQSDD14 pKa = 3.88PSSEE18 pKa = 4.02LLSFPLIFTNNDD30 pKa = 3.22SGSKK34 pKa = 8.52TLQPQVRR41 pKa = 11.84FSQLGDD47 pKa = 3.2IQGGKK52 pKa = 10.26DD53 pKa = 3.05EE54 pKa = 4.75TVFITLYY61 pKa = 11.1GFIQTEE67 pKa = 4.09QDD69 pKa = 4.11SLDD72 pKa = 3.63LTHH75 pKa = 6.4NQSTFQVLGQTSKK88 pKa = 11.11PDD90 pKa = 3.88TITAACIPLGATTLRR105 pKa = 11.84STITRR110 pKa = 11.84MAEE113 pKa = 3.68EE114 pKa = 4.12SLEE117 pKa = 4.02LLILVKK123 pKa = 10.51KK124 pKa = 10.42SAVSSEE130 pKa = 3.73KK131 pKa = 10.68LAVSFINIPPSLAGILVIRR150 pKa = 11.84AGGFILSAEE159 pKa = 4.6EE160 pKa = 4.22YY161 pKa = 10.08VKK163 pKa = 11.03SPSKK167 pKa = 10.63LQAGYY172 pKa = 9.62QYY174 pKa = 11.13KK175 pKa = 10.17FKK177 pKa = 10.59PVFITCTRR185 pKa = 11.84IFKK188 pKa = 10.58GKK190 pKa = 9.8LYY192 pKa = 10.26KK193 pKa = 10.29VPKK196 pKa = 8.6SMHH199 pKa = 6.19YY200 pKa = 9.7IASEE204 pKa = 3.99LLYY207 pKa = 10.79KK208 pKa = 10.57AVLEE212 pKa = 4.33IQFQLDD218 pKa = 3.73IKK220 pKa = 10.24PDD222 pKa = 3.49HH223 pKa = 6.98PQTKK227 pKa = 8.63MLKK230 pKa = 10.11KK231 pKa = 9.99QDD233 pKa = 3.75TNEE236 pKa = 3.76GPEE239 pKa = 4.01YY240 pKa = 10.83YY241 pKa = 10.79GFVWFHH247 pKa = 6.5LLNFKK252 pKa = 9.06KK253 pKa = 7.4TTARR257 pKa = 11.84GEE259 pKa = 3.75IRR261 pKa = 11.84TLEE264 pKa = 4.39KK265 pKa = 10.51ISDD268 pKa = 4.2KK269 pKa = 10.51IRR271 pKa = 11.84AMGIKK276 pKa = 10.13VSLYY280 pKa = 10.38DD281 pKa = 3.25LWGPTILAEE290 pKa = 4.02ITGKK294 pKa = 9.9KK295 pKa = 9.38SKK297 pKa = 9.98YY298 pKa = 8.52AQGFFSLNGCACLPVARR315 pKa = 11.84ASPEE319 pKa = 3.55IAKK322 pKa = 9.59LIWSCSTRR330 pKa = 11.84IKK332 pKa = 10.52SATIIVQSSDD342 pKa = 2.76KK343 pKa = 10.86RR344 pKa = 11.84GLLNSEE350 pKa = 4.11DD351 pKa = 4.61LEE353 pKa = 4.47IKK355 pKa = 10.34GAVGVSPRR363 pKa = 11.84KK364 pKa = 9.76LGSYY368 pKa = 10.68SLFKK372 pKa = 10.72KK373 pKa = 9.92HH374 pKa = 6.4
MM1 pKa = 7.85ASTSIPLHH9 pKa = 6.37INQSDD14 pKa = 3.88PSSEE18 pKa = 4.02LLSFPLIFTNNDD30 pKa = 3.22SGSKK34 pKa = 8.52TLQPQVRR41 pKa = 11.84FSQLGDD47 pKa = 3.2IQGGKK52 pKa = 10.26DD53 pKa = 3.05EE54 pKa = 4.75TVFITLYY61 pKa = 11.1GFIQTEE67 pKa = 4.09QDD69 pKa = 4.11SLDD72 pKa = 3.63LTHH75 pKa = 6.4NQSTFQVLGQTSKK88 pKa = 11.11PDD90 pKa = 3.88TITAACIPLGATTLRR105 pKa = 11.84STITRR110 pKa = 11.84MAEE113 pKa = 3.68EE114 pKa = 4.12SLEE117 pKa = 4.02LLILVKK123 pKa = 10.51KK124 pKa = 10.42SAVSSEE130 pKa = 3.73KK131 pKa = 10.68LAVSFINIPPSLAGILVIRR150 pKa = 11.84AGGFILSAEE159 pKa = 4.6EE160 pKa = 4.22YY161 pKa = 10.08VKK163 pKa = 11.03SPSKK167 pKa = 10.63LQAGYY172 pKa = 9.62QYY174 pKa = 11.13KK175 pKa = 10.17FKK177 pKa = 10.59PVFITCTRR185 pKa = 11.84IFKK188 pKa = 10.58GKK190 pKa = 9.8LYY192 pKa = 10.26KK193 pKa = 10.29VPKK196 pKa = 8.6SMHH199 pKa = 6.19YY200 pKa = 9.7IASEE204 pKa = 3.99LLYY207 pKa = 10.79KK208 pKa = 10.57AVLEE212 pKa = 4.33IQFQLDD218 pKa = 3.73IKK220 pKa = 10.24PDD222 pKa = 3.49HH223 pKa = 6.98PQTKK227 pKa = 8.63MLKK230 pKa = 10.11KK231 pKa = 9.99QDD233 pKa = 3.75TNEE236 pKa = 3.76GPEE239 pKa = 4.01YY240 pKa = 10.83YY241 pKa = 10.79GFVWFHH247 pKa = 6.5LLNFKK252 pKa = 9.06KK253 pKa = 7.4TTARR257 pKa = 11.84GEE259 pKa = 3.75IRR261 pKa = 11.84TLEE264 pKa = 4.39KK265 pKa = 10.51ISDD268 pKa = 4.2KK269 pKa = 10.51IRR271 pKa = 11.84AMGIKK276 pKa = 10.13VSLYY280 pKa = 10.38DD281 pKa = 3.25LWGPTILAEE290 pKa = 4.02ITGKK294 pKa = 9.9KK295 pKa = 9.38SKK297 pKa = 9.98YY298 pKa = 8.52AQGFFSLNGCACLPVARR315 pKa = 11.84ASPEE319 pKa = 3.55IAKK322 pKa = 9.59LIWSCSTRR330 pKa = 11.84IKK332 pKa = 10.52SATIIVQSSDD342 pKa = 2.76KK343 pKa = 10.86RR344 pKa = 11.84GLLNSEE350 pKa = 4.11DD351 pKa = 4.61LEE353 pKa = 4.47IKK355 pKa = 10.34GAVGVSPRR363 pKa = 11.84KK364 pKa = 9.76LGSYY368 pKa = 10.68SLFKK372 pKa = 10.72KK373 pKa = 9.92HH374 pKa = 6.4
Molecular weight: 41.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4581 |
374 |
2216 |
763.5 |
85.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.112 ± 0.611 | 1.877 ± 0.234 |
4.824 ± 0.359 | 5.523 ± 0.35 |
3.209 ± 0.377 | 5.152 ± 0.337 |
1.877 ± 0.333 | 8.557 ± 0.567 |
5.283 ± 0.552 | 11.111 ± 0.477 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.877 ± 0.286 | 5.108 ± 0.44 |
4.475 ± 0.499 | 4.431 ± 0.429 |
4.671 ± 0.533 | 9.146 ± 0.179 |
6.702 ± 0.406 | 5.763 ± 0.424 |
1.026 ± 0.187 | 3.274 ± 0.416 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
