
Aspergillus parasiticus (strain ATCC 56775 / NRRL 5862 / SRRC 143 / SU-1)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus; Aspergillus parasiticus
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
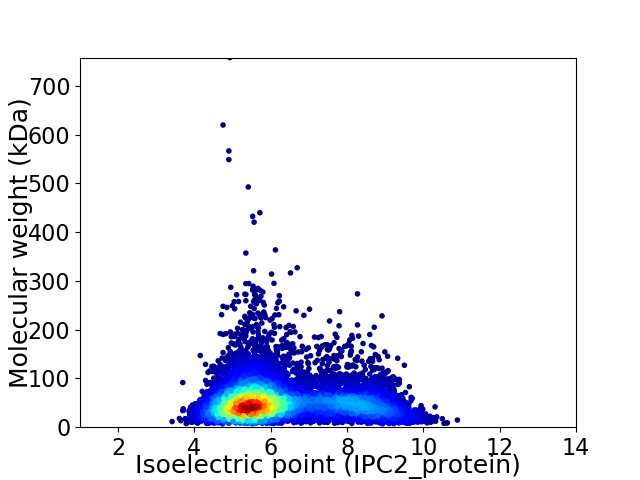
Virtual 2D-PAGE plot for 8651 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F0ICY2|A0A0F0ICY2_ASPPU DLH domain-containing protein OS=Aspergillus parasiticus (strain ATCC 56775 / NRRL 5862 / SRRC 143 / SU-1) OX=1403190 GN=P875_00010068 PE=4 SV=1
MM1 pKa = 7.7ADD3 pKa = 3.51EE4 pKa = 5.26AGSIYY9 pKa = 10.71DD10 pKa = 3.85EE11 pKa = 4.95IEE13 pKa = 4.34IEE15 pKa = 5.79DD16 pKa = 3.79MTFDD20 pKa = 6.24PITQLYY26 pKa = 9.73HH27 pKa = 5.63YY28 pKa = 7.55PCPCGDD34 pKa = 3.67RR35 pKa = 11.84FEE37 pKa = 5.7IMIDD41 pKa = 3.49DD42 pKa = 4.19LRR44 pKa = 11.84DD45 pKa = 3.58GEE47 pKa = 4.73EE48 pKa = 3.84IAVCPSCSLKK58 pKa = 10.28IRR60 pKa = 11.84VIFDD64 pKa = 3.02VDD66 pKa = 4.13DD67 pKa = 3.69LHH69 pKa = 9.25KK70 pKa = 10.81DD71 pKa = 3.63DD72 pKa = 4.29QQQGPSAVAVQAA84 pKa = 4.09
MM1 pKa = 7.7ADD3 pKa = 3.51EE4 pKa = 5.26AGSIYY9 pKa = 10.71DD10 pKa = 3.85EE11 pKa = 4.95IEE13 pKa = 4.34IEE15 pKa = 5.79DD16 pKa = 3.79MTFDD20 pKa = 6.24PITQLYY26 pKa = 9.73HH27 pKa = 5.63YY28 pKa = 7.55PCPCGDD34 pKa = 3.67RR35 pKa = 11.84FEE37 pKa = 5.7IMIDD41 pKa = 3.49DD42 pKa = 4.19LRR44 pKa = 11.84DD45 pKa = 3.58GEE47 pKa = 4.73EE48 pKa = 3.84IAVCPSCSLKK58 pKa = 10.28IRR60 pKa = 11.84VIFDD64 pKa = 3.02VDD66 pKa = 4.13DD67 pKa = 3.69LHH69 pKa = 9.25KK70 pKa = 10.81DD71 pKa = 3.63DD72 pKa = 4.29QQQGPSAVAVQAA84 pKa = 4.09
Molecular weight: 9.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F0IC83|A0A0F0IC83_ASPPU MATAHMG-box OS=Aspergillus parasiticus (strain ATCC 56775 / NRRL 5862 / SRRC 143 / SU-1) OX=1403190 GN=P875_00010325 PE=4 SV=1
MM1 pKa = 7.14EE2 pKa = 4.95QVSQPDD8 pKa = 3.49QMNHH12 pKa = 5.02FQHH15 pKa = 6.92LNRR18 pKa = 11.84IALHH22 pKa = 5.49EE23 pKa = 4.22FVRR26 pKa = 11.84LPVSRR31 pKa = 11.84EE32 pKa = 4.03MVAHH36 pKa = 7.41LAMQASQVIRR46 pKa = 11.84CEE48 pKa = 3.89PHH50 pKa = 4.85VTTASAHH57 pKa = 5.34GQPTPPSTPPLDD69 pKa = 4.68AVDD72 pKa = 4.09SQLPPLPSVEE82 pKa = 4.1MFITSLVTRR91 pKa = 11.84SQVQVPTLMTSLVYY105 pKa = 10.42LARR108 pKa = 11.84LRR110 pKa = 11.84ARR112 pKa = 11.84LPPVAKK118 pKa = 10.25GMRR121 pKa = 11.84CTVHH125 pKa = 7.16RR126 pKa = 11.84IFLASLILAAKK137 pKa = 9.3NLNDD141 pKa = 3.64SSPKK145 pKa = 7.95NKK147 pKa = 9.25HH148 pKa = 3.66WARR151 pKa = 11.84YY152 pKa = 4.24TTVKK156 pKa = 10.64GYY158 pKa = 10.95DD159 pKa = 3.2GFAFSLPEE167 pKa = 3.85VNLMEE172 pKa = 4.37RR173 pKa = 11.84QLLFLLDD180 pKa = 3.01WDD182 pKa = 4.16TRR184 pKa = 11.84VTEE187 pKa = 4.12EE188 pKa = 5.79DD189 pKa = 4.18LLCHH193 pKa = 7.13LEE195 pKa = 4.36PFLAPIRR202 pKa = 11.84YY203 pKa = 8.88RR204 pKa = 11.84YY205 pKa = 8.93QVQEE209 pKa = 4.06RR210 pKa = 11.84EE211 pKa = 4.18AEE213 pKa = 3.97LRR215 pKa = 11.84QPRR218 pKa = 11.84EE219 pKa = 3.57WRR221 pKa = 11.84RR222 pKa = 11.84LQASAEE228 pKa = 3.98LLACRR233 pKa = 11.84LRR235 pKa = 11.84RR236 pKa = 11.84QKK238 pKa = 11.1LEE240 pKa = 3.57ARR242 pKa = 11.84LEE244 pKa = 3.93ARR246 pKa = 11.84RR247 pKa = 11.84SDD249 pKa = 3.88SIHH252 pKa = 6.1RR253 pKa = 11.84RR254 pKa = 11.84QRR256 pKa = 11.84LPASPASSVSLSSMSSNASPASLADD281 pKa = 3.57TDD283 pKa = 3.38RR284 pKa = 11.84HH285 pKa = 5.3KK286 pKa = 10.59PYY288 pKa = 10.35RR289 pKa = 11.84PRR291 pKa = 11.84RR292 pKa = 11.84RR293 pKa = 11.84PSTRR297 pKa = 11.84SGASVSPPSAQDD309 pKa = 3.39VPSLTRR315 pKa = 11.84VDD317 pKa = 4.03TVPSLSSRR325 pKa = 11.84ASSIAPSSRR334 pKa = 11.84NATPASLRR342 pKa = 11.84TSSSITSMEE351 pKa = 3.45ADD353 pKa = 2.84IRR355 pKa = 11.84VVDD358 pKa = 4.64GARR361 pKa = 11.84SPSLSCGYY369 pKa = 10.53VPVSSMMAPNPKK381 pKa = 8.92MEE383 pKa = 5.64EE384 pKa = 3.74IQQPTKK390 pKa = 10.35KK391 pKa = 10.35VRR393 pKa = 11.84TNGHH397 pKa = 6.25AGHH400 pKa = 6.84TGFVARR406 pKa = 11.84FLASAAGSYY415 pKa = 9.41MGGRR419 pKa = 11.84MRR421 pKa = 11.84SHH423 pKa = 5.9VV424 pKa = 3.46
MM1 pKa = 7.14EE2 pKa = 4.95QVSQPDD8 pKa = 3.49QMNHH12 pKa = 5.02FQHH15 pKa = 6.92LNRR18 pKa = 11.84IALHH22 pKa = 5.49EE23 pKa = 4.22FVRR26 pKa = 11.84LPVSRR31 pKa = 11.84EE32 pKa = 4.03MVAHH36 pKa = 7.41LAMQASQVIRR46 pKa = 11.84CEE48 pKa = 3.89PHH50 pKa = 4.85VTTASAHH57 pKa = 5.34GQPTPPSTPPLDD69 pKa = 4.68AVDD72 pKa = 4.09SQLPPLPSVEE82 pKa = 4.1MFITSLVTRR91 pKa = 11.84SQVQVPTLMTSLVYY105 pKa = 10.42LARR108 pKa = 11.84LRR110 pKa = 11.84ARR112 pKa = 11.84LPPVAKK118 pKa = 10.25GMRR121 pKa = 11.84CTVHH125 pKa = 7.16RR126 pKa = 11.84IFLASLILAAKK137 pKa = 9.3NLNDD141 pKa = 3.64SSPKK145 pKa = 7.95NKK147 pKa = 9.25HH148 pKa = 3.66WARR151 pKa = 11.84YY152 pKa = 4.24TTVKK156 pKa = 10.64GYY158 pKa = 10.95DD159 pKa = 3.2GFAFSLPEE167 pKa = 3.85VNLMEE172 pKa = 4.37RR173 pKa = 11.84QLLFLLDD180 pKa = 3.01WDD182 pKa = 4.16TRR184 pKa = 11.84VTEE187 pKa = 4.12EE188 pKa = 5.79DD189 pKa = 4.18LLCHH193 pKa = 7.13LEE195 pKa = 4.36PFLAPIRR202 pKa = 11.84YY203 pKa = 8.88RR204 pKa = 11.84YY205 pKa = 8.93QVQEE209 pKa = 4.06RR210 pKa = 11.84EE211 pKa = 4.18AEE213 pKa = 3.97LRR215 pKa = 11.84QPRR218 pKa = 11.84EE219 pKa = 3.57WRR221 pKa = 11.84RR222 pKa = 11.84LQASAEE228 pKa = 3.98LLACRR233 pKa = 11.84LRR235 pKa = 11.84RR236 pKa = 11.84QKK238 pKa = 11.1LEE240 pKa = 3.57ARR242 pKa = 11.84LEE244 pKa = 3.93ARR246 pKa = 11.84RR247 pKa = 11.84SDD249 pKa = 3.88SIHH252 pKa = 6.1RR253 pKa = 11.84RR254 pKa = 11.84QRR256 pKa = 11.84LPASPASSVSLSSMSSNASPASLADD281 pKa = 3.57TDD283 pKa = 3.38RR284 pKa = 11.84HH285 pKa = 5.3KK286 pKa = 10.59PYY288 pKa = 10.35RR289 pKa = 11.84PRR291 pKa = 11.84RR292 pKa = 11.84RR293 pKa = 11.84PSTRR297 pKa = 11.84SGASVSPPSAQDD309 pKa = 3.39VPSLTRR315 pKa = 11.84VDD317 pKa = 4.03TVPSLSSRR325 pKa = 11.84ASSIAPSSRR334 pKa = 11.84NATPASLRR342 pKa = 11.84TSSSITSMEE351 pKa = 3.45ADD353 pKa = 2.84IRR355 pKa = 11.84VVDD358 pKa = 4.64GARR361 pKa = 11.84SPSLSCGYY369 pKa = 10.53VPVSSMMAPNPKK381 pKa = 8.92MEE383 pKa = 5.64EE384 pKa = 3.74IQQPTKK390 pKa = 10.35KK391 pKa = 10.35VRR393 pKa = 11.84TNGHH397 pKa = 6.25AGHH400 pKa = 6.84TGFVARR406 pKa = 11.84FLASAAGSYY415 pKa = 9.41MGGRR419 pKa = 11.84MRR421 pKa = 11.84SHH423 pKa = 5.9VV424 pKa = 3.46
Molecular weight: 47.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4430027 |
66 |
6885 |
512.1 |
56.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.328 ± 0.021 | 1.263 ± 0.009 |
5.62 ± 0.017 | 6.118 ± 0.02 |
3.832 ± 0.014 | 6.881 ± 0.022 |
2.413 ± 0.011 | 5.15 ± 0.017 |
4.636 ± 0.021 | 9.163 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.168 ± 0.009 | 3.748 ± 0.012 |
5.833 ± 0.027 | 3.999 ± 0.016 |
5.893 ± 0.02 | 8.199 ± 0.029 |
5.959 ± 0.016 | 6.307 ± 0.018 |
1.522 ± 0.009 | 2.965 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
