
Flavobacteriaceae bacterium M625
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; unclassified Flavobacteriaceae
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
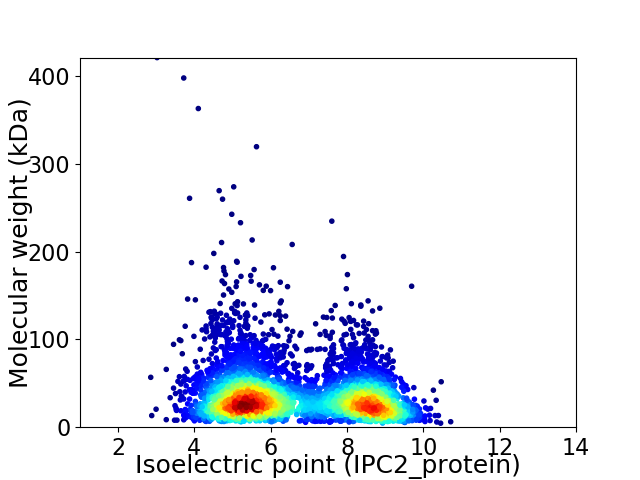
Virtual 2D-PAGE plot for 5099 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A504J552|A0A504J552_9FLAO Efflux RND transporter periplasmic adaptor subunit OS=Flavobacteriaceae bacterium M625 OX=2589995 GN=FHK87_12170 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.39KK3 pKa = 8.27ITRR6 pKa = 11.84VLSGFKK12 pKa = 10.05MPVLLFLFSFFSTFGQTGITSLSASYY38 pKa = 11.14KK39 pKa = 10.53DD40 pKa = 4.42INDD43 pKa = 3.58SSNDD47 pKa = 3.23FSNVGNGTNPGYY59 pKa = 10.94SSATTYY65 pKa = 11.61NLLFSTNTTIEE76 pKa = 3.73NDD78 pKa = 3.46YY79 pKa = 10.28TIDD82 pKa = 3.54GSFVAGGTTYY92 pKa = 9.93NTVILIDD99 pKa = 3.48QFEE102 pKa = 4.18VRR104 pKa = 11.84RR105 pKa = 11.84NGTIDD110 pKa = 3.45GQSNRR115 pKa = 11.84QILWFEE121 pKa = 4.28RR122 pKa = 11.84EE123 pKa = 4.38STSGTTINLRR133 pKa = 11.84PSYY136 pKa = 9.54VASVEE141 pKa = 3.88EE142 pKa = 4.23SFRR145 pKa = 11.84GFRR148 pKa = 11.84LNIGSDD154 pKa = 3.1NTFVNTGSNQSNNIEE169 pKa = 3.88RR170 pKa = 11.84FDD172 pKa = 4.52YY173 pKa = 10.98INSSPLSTNTTANAGFAIFEE193 pKa = 4.25RR194 pKa = 11.84DD195 pKa = 3.39GNDD198 pKa = 2.97SFKK201 pKa = 10.65IAAITGLDD209 pKa = 3.33AFGEE213 pKa = 4.48PNAFTPLLTVNTAAYY228 pKa = 9.58GPSLKK233 pKa = 10.1QVNYY237 pKa = 10.18TIFQKK242 pKa = 10.4QDD244 pKa = 3.01SEE246 pKa = 5.41SNLKK250 pKa = 9.44PAEE253 pKa = 4.22NGGPQQIRR261 pKa = 11.84GTFVSFQDD269 pKa = 3.82LGVIPNQAVYY279 pKa = 10.27GYY281 pKa = 10.72VILPADD287 pKa = 3.77ATSVDD292 pKa = 4.36FTTNPTNTDD301 pKa = 3.07SSNGGMDD308 pKa = 4.25AFPGGGFFDD317 pKa = 3.9SDD319 pKa = 3.34GGLTVVNMPDD329 pKa = 3.38ADD331 pKa = 3.88NDD333 pKa = 4.35GVSNTLDD340 pKa = 3.37QDD342 pKa = 3.84DD343 pKa = 5.19DD344 pKa = 4.34NDD346 pKa = 4.97GILDD350 pKa = 3.62INEE353 pKa = 4.53GLDD356 pKa = 3.81CGNGDD361 pKa = 3.16WTAGGTYY368 pKa = 10.3NINQNLGATINNTATYY384 pKa = 7.53NTRR387 pKa = 11.84DD388 pKa = 3.21VDD390 pKa = 4.0LTATLSGGATLSFIRR405 pKa = 11.84IQNAVGGNGQGIVQRR420 pKa = 11.84AQNSNFNTSPVDD432 pKa = 3.36YY433 pKa = 8.94TYY435 pKa = 11.15TFNDD439 pKa = 3.66PVVNLAFDD447 pKa = 4.92FGGLDD452 pKa = 3.18NNDD455 pKa = 3.45FVEE458 pKa = 4.51VRR460 pKa = 11.84AFLGANEE467 pKa = 4.99IILGPDD473 pKa = 2.9NFSVYY478 pKa = 9.75VASDD482 pKa = 2.87VSYY485 pKa = 11.3NGTSSFLATNSGAGNNSNNGSFSVEE510 pKa = 3.41IGDD513 pKa = 4.39PIDD516 pKa = 5.26RR517 pKa = 11.84IVLKK521 pKa = 9.58TGKK524 pKa = 8.36STGSGQVTLHH534 pKa = 6.14TSAFRR539 pKa = 11.84YY540 pKa = 8.45CIPRR544 pKa = 11.84DD545 pKa = 3.28TDD547 pKa = 3.32NNGVYY552 pKa = 10.6DD553 pKa = 4.64HH554 pKa = 7.63LDD556 pKa = 3.42TDD558 pKa = 4.35SDD560 pKa = 4.06NDD562 pKa = 3.66GCADD566 pKa = 4.59AIEE569 pKa = 4.78GGYY572 pKa = 8.72TDD574 pKa = 5.15PDD576 pKa = 3.72DD577 pKa = 6.73DD578 pKa = 5.32EE579 pKa = 5.97ILGNSPVSVDD589 pKa = 3.66GIGRR593 pKa = 11.84VTGQGGYY600 pKa = 9.78SGTRR604 pKa = 11.84SGVITFGTLPSITAQSANQTINTGSNTSFTVTATGSDD641 pKa = 3.33LTYY644 pKa = 10.32RR645 pKa = 11.84WQVDD649 pKa = 3.34TGGGFVDD656 pKa = 4.34IDD658 pKa = 3.77PANTTDD664 pKa = 4.01IYY666 pKa = 10.56TGSDD670 pKa = 3.18TASLNLTNVPVSDD683 pKa = 4.54DD684 pKa = 3.54GNLYY688 pKa = 10.21RR689 pKa = 11.84VIVSSTSFMCGNVEE703 pKa = 4.42SSSILLNVNNYY714 pKa = 7.22TVNLTASVDD723 pKa = 3.74TAVEE727 pKa = 4.03GGIDD731 pKa = 3.88GEE733 pKa = 4.69FTATLNAVNTTGSAIVIPVVMSGTATDD760 pKa = 3.82GTDD763 pKa = 3.06YY764 pKa = 11.21TSIVNITIPNGSDD777 pKa = 2.74TGTVAVDD784 pKa = 3.25AATDD788 pKa = 3.77GLIEE792 pKa = 4.36GDD794 pKa = 3.45EE795 pKa = 4.14TAIASLGTLPSGVVAGTSTSDD816 pKa = 2.88TVTISDD822 pKa = 3.42ATEE825 pKa = 3.79YY826 pKa = 10.07TVNLTASVDD835 pKa = 3.76TAVEE839 pKa = 4.29GGTDD843 pKa = 3.94GEE845 pKa = 4.73FTATLNAVNTTGSAIVIPVVMSGTATDD872 pKa = 3.82GTDD875 pKa = 3.06YY876 pKa = 11.21TSIVNITIPNGSDD889 pKa = 2.74TGTVAVDD896 pKa = 3.25AATDD900 pKa = 3.77GLIEE904 pKa = 4.36GDD906 pKa = 3.37EE907 pKa = 4.14TAIARR912 pKa = 11.84WRR914 pKa = 11.84VYY916 pKa = 11.31SDD918 pKa = 3.86FKK920 pKa = 11.05CGEE923 pKa = 4.4HH924 pKa = 6.71YY925 pKa = 10.13RR926 pKa = 11.84QCYY929 pKa = 9.6CDD931 pKa = 3.59PCSDD935 pKa = 3.01EE936 pKa = 4.25WYY938 pKa = 10.61GYY940 pKa = 10.84
MM1 pKa = 7.44KK2 pKa = 10.39KK3 pKa = 8.27ITRR6 pKa = 11.84VLSGFKK12 pKa = 10.05MPVLLFLFSFFSTFGQTGITSLSASYY38 pKa = 11.14KK39 pKa = 10.53DD40 pKa = 4.42INDD43 pKa = 3.58SSNDD47 pKa = 3.23FSNVGNGTNPGYY59 pKa = 10.94SSATTYY65 pKa = 11.61NLLFSTNTTIEE76 pKa = 3.73NDD78 pKa = 3.46YY79 pKa = 10.28TIDD82 pKa = 3.54GSFVAGGTTYY92 pKa = 9.93NTVILIDD99 pKa = 3.48QFEE102 pKa = 4.18VRR104 pKa = 11.84RR105 pKa = 11.84NGTIDD110 pKa = 3.45GQSNRR115 pKa = 11.84QILWFEE121 pKa = 4.28RR122 pKa = 11.84EE123 pKa = 4.38STSGTTINLRR133 pKa = 11.84PSYY136 pKa = 9.54VASVEE141 pKa = 3.88EE142 pKa = 4.23SFRR145 pKa = 11.84GFRR148 pKa = 11.84LNIGSDD154 pKa = 3.1NTFVNTGSNQSNNIEE169 pKa = 3.88RR170 pKa = 11.84FDD172 pKa = 4.52YY173 pKa = 10.98INSSPLSTNTTANAGFAIFEE193 pKa = 4.25RR194 pKa = 11.84DD195 pKa = 3.39GNDD198 pKa = 2.97SFKK201 pKa = 10.65IAAITGLDD209 pKa = 3.33AFGEE213 pKa = 4.48PNAFTPLLTVNTAAYY228 pKa = 9.58GPSLKK233 pKa = 10.1QVNYY237 pKa = 10.18TIFQKK242 pKa = 10.4QDD244 pKa = 3.01SEE246 pKa = 5.41SNLKK250 pKa = 9.44PAEE253 pKa = 4.22NGGPQQIRR261 pKa = 11.84GTFVSFQDD269 pKa = 3.82LGVIPNQAVYY279 pKa = 10.27GYY281 pKa = 10.72VILPADD287 pKa = 3.77ATSVDD292 pKa = 4.36FTTNPTNTDD301 pKa = 3.07SSNGGMDD308 pKa = 4.25AFPGGGFFDD317 pKa = 3.9SDD319 pKa = 3.34GGLTVVNMPDD329 pKa = 3.38ADD331 pKa = 3.88NDD333 pKa = 4.35GVSNTLDD340 pKa = 3.37QDD342 pKa = 3.84DD343 pKa = 5.19DD344 pKa = 4.34NDD346 pKa = 4.97GILDD350 pKa = 3.62INEE353 pKa = 4.53GLDD356 pKa = 3.81CGNGDD361 pKa = 3.16WTAGGTYY368 pKa = 10.3NINQNLGATINNTATYY384 pKa = 7.53NTRR387 pKa = 11.84DD388 pKa = 3.21VDD390 pKa = 4.0LTATLSGGATLSFIRR405 pKa = 11.84IQNAVGGNGQGIVQRR420 pKa = 11.84AQNSNFNTSPVDD432 pKa = 3.36YY433 pKa = 8.94TYY435 pKa = 11.15TFNDD439 pKa = 3.66PVVNLAFDD447 pKa = 4.92FGGLDD452 pKa = 3.18NNDD455 pKa = 3.45FVEE458 pKa = 4.51VRR460 pKa = 11.84AFLGANEE467 pKa = 4.99IILGPDD473 pKa = 2.9NFSVYY478 pKa = 9.75VASDD482 pKa = 2.87VSYY485 pKa = 11.3NGTSSFLATNSGAGNNSNNGSFSVEE510 pKa = 3.41IGDD513 pKa = 4.39PIDD516 pKa = 5.26RR517 pKa = 11.84IVLKK521 pKa = 9.58TGKK524 pKa = 8.36STGSGQVTLHH534 pKa = 6.14TSAFRR539 pKa = 11.84YY540 pKa = 8.45CIPRR544 pKa = 11.84DD545 pKa = 3.28TDD547 pKa = 3.32NNGVYY552 pKa = 10.6DD553 pKa = 4.64HH554 pKa = 7.63LDD556 pKa = 3.42TDD558 pKa = 4.35SDD560 pKa = 4.06NDD562 pKa = 3.66GCADD566 pKa = 4.59AIEE569 pKa = 4.78GGYY572 pKa = 8.72TDD574 pKa = 5.15PDD576 pKa = 3.72DD577 pKa = 6.73DD578 pKa = 5.32EE579 pKa = 5.97ILGNSPVSVDD589 pKa = 3.66GIGRR593 pKa = 11.84VTGQGGYY600 pKa = 9.78SGTRR604 pKa = 11.84SGVITFGTLPSITAQSANQTINTGSNTSFTVTATGSDD641 pKa = 3.33LTYY644 pKa = 10.32RR645 pKa = 11.84WQVDD649 pKa = 3.34TGGGFVDD656 pKa = 4.34IDD658 pKa = 3.77PANTTDD664 pKa = 4.01IYY666 pKa = 10.56TGSDD670 pKa = 3.18TASLNLTNVPVSDD683 pKa = 4.54DD684 pKa = 3.54GNLYY688 pKa = 10.21RR689 pKa = 11.84VIVSSTSFMCGNVEE703 pKa = 4.42SSSILLNVNNYY714 pKa = 7.22TVNLTASVDD723 pKa = 3.74TAVEE727 pKa = 4.03GGIDD731 pKa = 3.88GEE733 pKa = 4.69FTATLNAVNTTGSAIVIPVVMSGTATDD760 pKa = 3.82GTDD763 pKa = 3.06YY764 pKa = 11.21TSIVNITIPNGSDD777 pKa = 2.74TGTVAVDD784 pKa = 3.25AATDD788 pKa = 3.77GLIEE792 pKa = 4.36GDD794 pKa = 3.45EE795 pKa = 4.14TAIASLGTLPSGVVAGTSTSDD816 pKa = 2.88TVTISDD822 pKa = 3.42ATEE825 pKa = 3.79YY826 pKa = 10.07TVNLTASVDD835 pKa = 3.76TAVEE839 pKa = 4.29GGTDD843 pKa = 3.94GEE845 pKa = 4.73FTATLNAVNTTGSAIVIPVVMSGTATDD872 pKa = 3.82GTDD875 pKa = 3.06YY876 pKa = 11.21TSIVNITIPNGSDD889 pKa = 2.74TGTVAVDD896 pKa = 3.25AATDD900 pKa = 3.77GLIEE904 pKa = 4.36GDD906 pKa = 3.37EE907 pKa = 4.14TAIARR912 pKa = 11.84WRR914 pKa = 11.84VYY916 pKa = 11.31SDD918 pKa = 3.86FKK920 pKa = 11.05CGEE923 pKa = 4.4HH924 pKa = 6.71YY925 pKa = 10.13RR926 pKa = 11.84QCYY929 pKa = 9.6CDD931 pKa = 3.59PCSDD935 pKa = 3.01EE936 pKa = 4.25WYY938 pKa = 10.61GYY940 pKa = 10.84
Molecular weight: 98.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A504JA88|A0A504JA88_9FLAO Uncharacterized protein OS=Flavobacteriaceae bacterium M625 OX=2589995 GN=FHK87_09495 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 5.43KK51 pKa = 10.29HH52 pKa = 5.04
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 5.43KK51 pKa = 10.29HH52 pKa = 5.04
Molecular weight: 6.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1710689 |
38 |
4048 |
335.5 |
38.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.677 ± 0.029 | 0.774 ± 0.011 |
5.741 ± 0.028 | 6.702 ± 0.034 |
5.199 ± 0.026 | 6.02 ± 0.033 |
1.821 ± 0.018 | 8.634 ± 0.032 |
8.088 ± 0.05 | 9.012 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.99 ± 0.016 | 6.277 ± 0.034 |
3.32 ± 0.024 | 3.623 ± 0.02 |
3.476 ± 0.023 | 6.61 ± 0.03 |
5.971 ± 0.042 | 5.786 ± 0.027 |
1.07 ± 0.013 | 4.21 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
