
Maize streak virus genotype A (isolate South Africa) (MSV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus; Maize streak virus
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
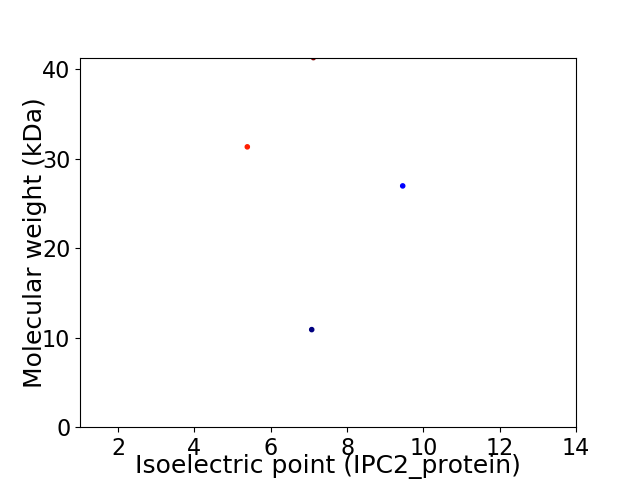
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
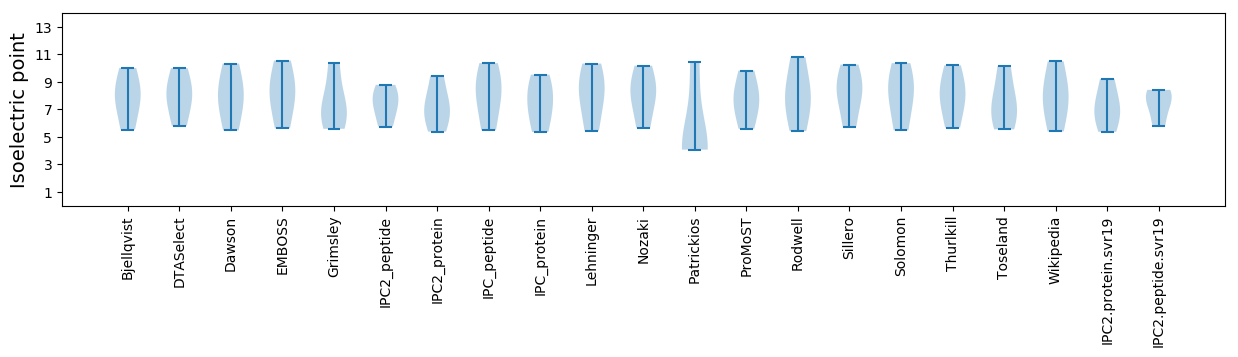
Protein with the lowest isoelectric point:
>sp|P14992|MP_MSVS Movement protein OS=Maize streak virus genotype A (isolate South Africa) OX=10824 GN=V2 PE=3 SV=1
MM1 pKa = 7.64ASSSSNRR8 pKa = 11.84QFSHH12 pKa = 7.0RR13 pKa = 11.84NANTFLTYY21 pKa = 9.4PKK23 pKa = 10.28CPEE26 pKa = 3.99NPEE29 pKa = 4.19IACQMIWEE37 pKa = 4.44LVVRR41 pKa = 11.84WIPKK45 pKa = 9.48YY46 pKa = 9.99ILCARR51 pKa = 11.84EE52 pKa = 3.78AHH54 pKa = 6.78KK55 pKa = 11.03DD56 pKa = 3.56GSLHH60 pKa = 6.61LHH62 pKa = 6.79ALLQTEE68 pKa = 4.27KK69 pKa = 10.2PVRR72 pKa = 11.84ISDD75 pKa = 3.96SRR77 pKa = 11.84FFDD80 pKa = 3.32INGFHH85 pKa = 7.46PNIQSAKK92 pKa = 8.27SVNRR96 pKa = 11.84VRR98 pKa = 11.84DD99 pKa = 3.85YY100 pKa = 11.14ILKK103 pKa = 10.0EE104 pKa = 3.56PLAVFEE110 pKa = 5.15RR111 pKa = 11.84GTFIPRR117 pKa = 11.84KK118 pKa = 9.9SPFLGKK124 pKa = 9.77SDD126 pKa = 4.69SEE128 pKa = 4.48VKK130 pKa = 9.84EE131 pKa = 4.11KK132 pKa = 10.86KK133 pKa = 9.48PSKK136 pKa = 10.42DD137 pKa = 2.99EE138 pKa = 3.71IMRR141 pKa = 11.84DD142 pKa = 3.68IISHH146 pKa = 5.71ATSKK150 pKa = 11.27AEE152 pKa = 3.98YY153 pKa = 10.23LSMIQKK159 pKa = 8.82EE160 pKa = 4.59LPFDD164 pKa = 3.51WSTKK168 pKa = 8.61LQYY171 pKa = 10.45FEE173 pKa = 4.94YY174 pKa = 10.26SANKK178 pKa = 9.59LFPEE182 pKa = 4.01IQEE185 pKa = 4.39EE186 pKa = 4.56FTNPHH191 pKa = 6.82PPSSPDD197 pKa = 3.42LLCNEE202 pKa = 5.57SINDD206 pKa = 3.68WLQPNIFQVSPEE218 pKa = 4.27AYY220 pKa = 9.18MLLQPTCYY228 pKa = 9.44TLEE231 pKa = 4.25DD232 pKa = 5.23AISDD236 pKa = 4.27LQWMDD241 pKa = 3.46SVSSHH246 pKa = 5.65QMKK249 pKa = 10.2DD250 pKa = 3.22QEE252 pKa = 4.62SRR254 pKa = 11.84ASTSSAQQEE263 pKa = 4.43PEE265 pKa = 3.87NLLGPEE271 pKa = 3.97AA272 pKa = 4.79
MM1 pKa = 7.64ASSSSNRR8 pKa = 11.84QFSHH12 pKa = 7.0RR13 pKa = 11.84NANTFLTYY21 pKa = 9.4PKK23 pKa = 10.28CPEE26 pKa = 3.99NPEE29 pKa = 4.19IACQMIWEE37 pKa = 4.44LVVRR41 pKa = 11.84WIPKK45 pKa = 9.48YY46 pKa = 9.99ILCARR51 pKa = 11.84EE52 pKa = 3.78AHH54 pKa = 6.78KK55 pKa = 11.03DD56 pKa = 3.56GSLHH60 pKa = 6.61LHH62 pKa = 6.79ALLQTEE68 pKa = 4.27KK69 pKa = 10.2PVRR72 pKa = 11.84ISDD75 pKa = 3.96SRR77 pKa = 11.84FFDD80 pKa = 3.32INGFHH85 pKa = 7.46PNIQSAKK92 pKa = 8.27SVNRR96 pKa = 11.84VRR98 pKa = 11.84DD99 pKa = 3.85YY100 pKa = 11.14ILKK103 pKa = 10.0EE104 pKa = 3.56PLAVFEE110 pKa = 5.15RR111 pKa = 11.84GTFIPRR117 pKa = 11.84KK118 pKa = 9.9SPFLGKK124 pKa = 9.77SDD126 pKa = 4.69SEE128 pKa = 4.48VKK130 pKa = 9.84EE131 pKa = 4.11KK132 pKa = 10.86KK133 pKa = 9.48PSKK136 pKa = 10.42DD137 pKa = 2.99EE138 pKa = 3.71IMRR141 pKa = 11.84DD142 pKa = 3.68IISHH146 pKa = 5.71ATSKK150 pKa = 11.27AEE152 pKa = 3.98YY153 pKa = 10.23LSMIQKK159 pKa = 8.82EE160 pKa = 4.59LPFDD164 pKa = 3.51WSTKK168 pKa = 8.61LQYY171 pKa = 10.45FEE173 pKa = 4.94YY174 pKa = 10.26SANKK178 pKa = 9.59LFPEE182 pKa = 4.01IQEE185 pKa = 4.39EE186 pKa = 4.56FTNPHH191 pKa = 6.82PPSSPDD197 pKa = 3.42LLCNEE202 pKa = 5.57SINDD206 pKa = 3.68WLQPNIFQVSPEE218 pKa = 4.27AYY220 pKa = 9.18MLLQPTCYY228 pKa = 9.44TLEE231 pKa = 4.25DD232 pKa = 5.23AISDD236 pKa = 4.27LQWMDD241 pKa = 3.46SVSSHH246 pKa = 5.65QMKK249 pKa = 10.2DD250 pKa = 3.22QEE252 pKa = 4.62SRR254 pKa = 11.84ASTSSAQQEE263 pKa = 4.43PEE265 pKa = 3.87NLLGPEE271 pKa = 3.97AA272 pKa = 4.79
Molecular weight: 31.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P14989|REP_MSVS Replication-associated protein OS=Maize streak virus genotype A (isolate South Africa) OX=10824 GN=C1/C2 PE=3 SV=2
MM1 pKa = 6.95STSKK5 pKa = 10.64RR6 pKa = 11.84KK7 pKa = 10.25RR8 pKa = 11.84GDD10 pKa = 3.18DD11 pKa = 3.77ANWSKK16 pKa = 10.82RR17 pKa = 11.84VPKK20 pKa = 10.12KK21 pKa = 10.5KK22 pKa = 9.93PSSAGLKK29 pKa = 9.76RR30 pKa = 11.84AGSKK34 pKa = 10.46ADD36 pKa = 3.86RR37 pKa = 11.84PSLQIQTLQHH47 pKa = 6.77AGTTMITVPSGGVCDD62 pKa = 6.17LINTYY67 pKa = 11.02ARR69 pKa = 11.84GSDD72 pKa = 3.04EE73 pKa = 4.39GNRR76 pKa = 11.84HH77 pKa = 4.54TSEE80 pKa = 3.76TLTYY84 pKa = 9.9KK85 pKa = 10.41IAVDD89 pKa = 3.29YY90 pKa = 11.37HH91 pKa = 6.12FVADD95 pKa = 4.52AAACRR100 pKa = 11.84YY101 pKa = 9.77SNTGTGVMWLVYY113 pKa = 9.18DD114 pKa = 4.16TTPGGQAPTPQTIFAYY130 pKa = 9.88PDD132 pKa = 3.36TLKK135 pKa = 10.77AWPATWKK142 pKa = 10.4VSRR145 pKa = 11.84EE146 pKa = 3.81LCHH149 pKa = 6.49RR150 pKa = 11.84FVVKK154 pKa = 10.43RR155 pKa = 11.84RR156 pKa = 11.84WLFNMEE162 pKa = 3.66TDD164 pKa = 3.45GRR166 pKa = 11.84IGSDD170 pKa = 2.88IPPSNTSWKK179 pKa = 7.81PCKK182 pKa = 9.99RR183 pKa = 11.84NIYY186 pKa = 7.92FHH188 pKa = 7.28KK189 pKa = 9.03FTSGLGVRR197 pKa = 11.84TQWKK201 pKa = 8.76NVTDD205 pKa = 4.02GGVGAIQRR213 pKa = 11.84GALYY217 pKa = 9.56MVIAPGNGLTFTAHH231 pKa = 5.61GQTRR235 pKa = 11.84LYY237 pKa = 9.73FKK239 pKa = 10.92SVGNQQ244 pKa = 2.8
MM1 pKa = 6.95STSKK5 pKa = 10.64RR6 pKa = 11.84KK7 pKa = 10.25RR8 pKa = 11.84GDD10 pKa = 3.18DD11 pKa = 3.77ANWSKK16 pKa = 10.82RR17 pKa = 11.84VPKK20 pKa = 10.12KK21 pKa = 10.5KK22 pKa = 9.93PSSAGLKK29 pKa = 9.76RR30 pKa = 11.84AGSKK34 pKa = 10.46ADD36 pKa = 3.86RR37 pKa = 11.84PSLQIQTLQHH47 pKa = 6.77AGTTMITVPSGGVCDD62 pKa = 6.17LINTYY67 pKa = 11.02ARR69 pKa = 11.84GSDD72 pKa = 3.04EE73 pKa = 4.39GNRR76 pKa = 11.84HH77 pKa = 4.54TSEE80 pKa = 3.76TLTYY84 pKa = 9.9KK85 pKa = 10.41IAVDD89 pKa = 3.29YY90 pKa = 11.37HH91 pKa = 6.12FVADD95 pKa = 4.52AAACRR100 pKa = 11.84YY101 pKa = 9.77SNTGTGVMWLVYY113 pKa = 9.18DD114 pKa = 4.16TTPGGQAPTPQTIFAYY130 pKa = 9.88PDD132 pKa = 3.36TLKK135 pKa = 10.77AWPATWKK142 pKa = 10.4VSRR145 pKa = 11.84EE146 pKa = 3.81LCHH149 pKa = 6.49RR150 pKa = 11.84FVVKK154 pKa = 10.43RR155 pKa = 11.84RR156 pKa = 11.84WLFNMEE162 pKa = 3.66TDD164 pKa = 3.45GRR166 pKa = 11.84IGSDD170 pKa = 2.88IPPSNTSWKK179 pKa = 7.81PCKK182 pKa = 9.99RR183 pKa = 11.84NIYY186 pKa = 7.92FHH188 pKa = 7.28KK189 pKa = 9.03FTSGLGVRR197 pKa = 11.84TQWKK201 pKa = 8.76NVTDD205 pKa = 4.02GGVGAIQRR213 pKa = 11.84GALYY217 pKa = 9.56MVIAPGNGLTFTAHH231 pKa = 5.61GQTRR235 pKa = 11.84LYY237 pKa = 9.73FKK239 pKa = 10.92SVGNQQ244 pKa = 2.8
Molecular weight: 26.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
972 |
101 |
355 |
243.0 |
27.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.967 ± 0.863 | 1.852 ± 0.214 |
4.835 ± 0.322 | 5.761 ± 1.434 |
4.321 ± 0.341 | 5.144 ± 1.782 |
2.263 ± 0.403 | 5.967 ± 0.718 |
6.687 ± 1.167 | 7.305 ± 1.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.058 ± 0.217 | 4.835 ± 0.384 |
7.716 ± 1.056 | 4.527 ± 0.376 |
5.247 ± 0.566 | 9.053 ± 0.945 |
5.35 ± 1.491 | 5.041 ± 0.817 |
2.469 ± 0.261 | 3.601 ± 0.231 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
