
Aquimarina atlantica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aquimarina
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
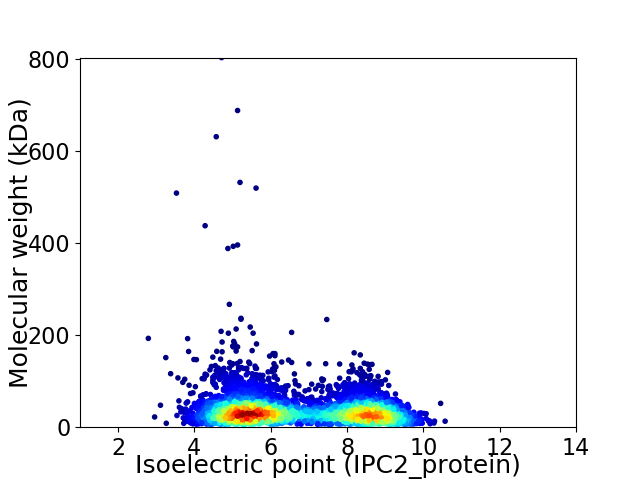
Virtual 2D-PAGE plot for 4786 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A023BS10|A0A023BS10_9FLAO Phosphatidate cytidylyltransferase OS=Aquimarina atlantica OX=1317122 GN=ATO12_21840 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 9.3MAKK5 pKa = 9.81GLPQILLLFSILVFKK20 pKa = 10.52STFFYY25 pKa = 10.92AQDD28 pKa = 3.23FDD30 pKa = 4.35VACRR34 pKa = 11.84AAQPFCSDD42 pKa = 3.29ASLEE46 pKa = 4.16LTFPNVTGDD55 pKa = 3.23MDD57 pKa = 4.08GLGEE61 pKa = 4.23VGCLNTTPNPAWYY74 pKa = 9.07FIRR77 pKa = 11.84IDD79 pKa = 3.58QPGEE83 pKa = 4.02LIFDD87 pKa = 3.96IQQWVDD93 pKa = 3.51SNDD96 pKa = 3.22NSNLDD101 pKa = 3.39RR102 pKa = 11.84GEE104 pKa = 4.05RR105 pKa = 11.84QLDD108 pKa = 3.2VDD110 pKa = 4.8FIAWGPFPTSFVNCDD125 pKa = 3.47ILSQGCDD132 pKa = 3.22NNGDD136 pKa = 3.76GDD138 pKa = 4.29NIRR141 pKa = 11.84PAEE144 pKa = 4.18CVNNVDD150 pKa = 5.09DD151 pKa = 4.84PDD153 pKa = 4.4YY154 pKa = 11.22YY155 pKa = 10.9VNNMDD160 pKa = 3.36NTNIIDD166 pKa = 3.53CSYY169 pKa = 11.53ARR171 pKa = 11.84TPDD174 pKa = 3.48DD175 pKa = 4.86DD176 pKa = 5.14IYY178 pKa = 10.94IEE180 pKa = 4.36TLTIPNAQSGEE191 pKa = 4.17YY192 pKa = 10.37YY193 pKa = 10.19IILITNFADD202 pKa = 3.61EE203 pKa = 4.92AGVIQLQQTNLNEE216 pKa = 4.62DD217 pKa = 3.38NAGTTDD223 pKa = 4.09CSILEE228 pKa = 4.35PGVGPDD234 pKa = 3.03IATCGEE240 pKa = 4.07FPITIEE246 pKa = 3.87GRR248 pKa = 11.84FPRR251 pKa = 11.84AVTYY255 pKa = 9.38QWARR259 pKa = 11.84ADD261 pKa = 3.41VGTTNFVNVPGPPGVIPFLEE281 pKa = 4.47VTTEE285 pKa = 3.7GVYY288 pKa = 8.85QLRR291 pKa = 11.84GFDD294 pKa = 3.37EE295 pKa = 4.73TGAEE299 pKa = 4.41VPDD302 pKa = 4.25SPDD305 pKa = 3.37DD306 pKa = 3.99LVVLDD311 pKa = 4.16VSDD314 pKa = 4.14AVVSVGYY321 pKa = 10.4SIAEE325 pKa = 3.75EE326 pKa = 4.39SFAGGYY332 pKa = 8.09TITAEE337 pKa = 4.24VVAAPDD343 pKa = 3.38VEE345 pKa = 4.53AAGFDD350 pKa = 3.66DD351 pKa = 5.22FEE353 pKa = 4.63YY354 pKa = 11.06RR355 pKa = 11.84LDD357 pKa = 3.83RR358 pKa = 11.84DD359 pKa = 3.11IGNGFLEE366 pKa = 4.0YY367 pKa = 10.54RR368 pKa = 11.84PFQSSPIFTNVPPGDD383 pKa = 3.72YY384 pKa = 10.54FITARR389 pKa = 11.84YY390 pKa = 8.34ANCPASEE397 pKa = 4.6EE398 pKa = 4.02ISDD401 pKa = 3.94VIMILGYY408 pKa = 9.72PKK410 pKa = 10.64YY411 pKa = 9.13FTPNGDD417 pKa = 3.65GFHH420 pKa = 6.88DD421 pKa = 3.78TWSLINIEE429 pKa = 4.6DD430 pKa = 3.67QPTALIYY437 pKa = 10.63IFDD440 pKa = 4.23RR441 pKa = 11.84YY442 pKa = 10.65GKK444 pKa = 10.0LLKK447 pKa = 9.8QLRR450 pKa = 11.84AGGPGWDD457 pKa = 3.18GTYY460 pKa = 10.4NGKK463 pKa = 9.74NMPSSDD469 pKa = 2.8YY470 pKa = 9.98WFRR473 pKa = 11.84VEE475 pKa = 4.92FNEE478 pKa = 4.68PRR480 pKa = 11.84DD481 pKa = 3.44PNMRR485 pKa = 11.84RR486 pKa = 11.84RR487 pKa = 11.84VFAGNFSLIRR497 pKa = 4.27
MM1 pKa = 7.47KK2 pKa = 9.3MAKK5 pKa = 9.81GLPQILLLFSILVFKK20 pKa = 10.52STFFYY25 pKa = 10.92AQDD28 pKa = 3.23FDD30 pKa = 4.35VACRR34 pKa = 11.84AAQPFCSDD42 pKa = 3.29ASLEE46 pKa = 4.16LTFPNVTGDD55 pKa = 3.23MDD57 pKa = 4.08GLGEE61 pKa = 4.23VGCLNTTPNPAWYY74 pKa = 9.07FIRR77 pKa = 11.84IDD79 pKa = 3.58QPGEE83 pKa = 4.02LIFDD87 pKa = 3.96IQQWVDD93 pKa = 3.51SNDD96 pKa = 3.22NSNLDD101 pKa = 3.39RR102 pKa = 11.84GEE104 pKa = 4.05RR105 pKa = 11.84QLDD108 pKa = 3.2VDD110 pKa = 4.8FIAWGPFPTSFVNCDD125 pKa = 3.47ILSQGCDD132 pKa = 3.22NNGDD136 pKa = 3.76GDD138 pKa = 4.29NIRR141 pKa = 11.84PAEE144 pKa = 4.18CVNNVDD150 pKa = 5.09DD151 pKa = 4.84PDD153 pKa = 4.4YY154 pKa = 11.22YY155 pKa = 10.9VNNMDD160 pKa = 3.36NTNIIDD166 pKa = 3.53CSYY169 pKa = 11.53ARR171 pKa = 11.84TPDD174 pKa = 3.48DD175 pKa = 4.86DD176 pKa = 5.14IYY178 pKa = 10.94IEE180 pKa = 4.36TLTIPNAQSGEE191 pKa = 4.17YY192 pKa = 10.37YY193 pKa = 10.19IILITNFADD202 pKa = 3.61EE203 pKa = 4.92AGVIQLQQTNLNEE216 pKa = 4.62DD217 pKa = 3.38NAGTTDD223 pKa = 4.09CSILEE228 pKa = 4.35PGVGPDD234 pKa = 3.03IATCGEE240 pKa = 4.07FPITIEE246 pKa = 3.87GRR248 pKa = 11.84FPRR251 pKa = 11.84AVTYY255 pKa = 9.38QWARR259 pKa = 11.84ADD261 pKa = 3.41VGTTNFVNVPGPPGVIPFLEE281 pKa = 4.47VTTEE285 pKa = 3.7GVYY288 pKa = 8.85QLRR291 pKa = 11.84GFDD294 pKa = 3.37EE295 pKa = 4.73TGAEE299 pKa = 4.41VPDD302 pKa = 4.25SPDD305 pKa = 3.37DD306 pKa = 3.99LVVLDD311 pKa = 4.16VSDD314 pKa = 4.14AVVSVGYY321 pKa = 10.4SIAEE325 pKa = 3.75EE326 pKa = 4.39SFAGGYY332 pKa = 8.09TITAEE337 pKa = 4.24VVAAPDD343 pKa = 3.38VEE345 pKa = 4.53AAGFDD350 pKa = 3.66DD351 pKa = 5.22FEE353 pKa = 4.63YY354 pKa = 11.06RR355 pKa = 11.84LDD357 pKa = 3.83RR358 pKa = 11.84DD359 pKa = 3.11IGNGFLEE366 pKa = 4.0YY367 pKa = 10.54RR368 pKa = 11.84PFQSSPIFTNVPPGDD383 pKa = 3.72YY384 pKa = 10.54FITARR389 pKa = 11.84YY390 pKa = 8.34ANCPASEE397 pKa = 4.6EE398 pKa = 4.02ISDD401 pKa = 3.94VIMILGYY408 pKa = 9.72PKK410 pKa = 10.64YY411 pKa = 9.13FTPNGDD417 pKa = 3.65GFHH420 pKa = 6.88DD421 pKa = 3.78TWSLINIEE429 pKa = 4.6DD430 pKa = 3.67QPTALIYY437 pKa = 10.63IFDD440 pKa = 4.23RR441 pKa = 11.84YY442 pKa = 10.65GKK444 pKa = 10.0LLKK447 pKa = 9.8QLRR450 pKa = 11.84AGGPGWDD457 pKa = 3.18GTYY460 pKa = 10.4NGKK463 pKa = 9.74NMPSSDD469 pKa = 2.8YY470 pKa = 9.98WFRR473 pKa = 11.84VEE475 pKa = 4.92FNEE478 pKa = 4.68PRR480 pKa = 11.84DD481 pKa = 3.44PNMRR485 pKa = 11.84RR486 pKa = 11.84RR487 pKa = 11.84VFAGNFSLIRR497 pKa = 4.27
Molecular weight: 55.17 kDa
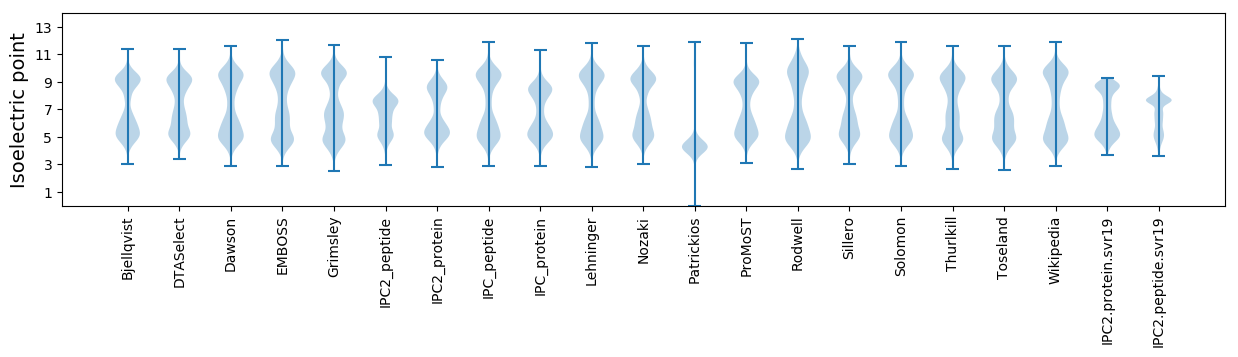
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A023BRY5|A0A023BRY5_9FLAO Uncharacterized protein OS=Aquimarina atlantica OX=1317122 GN=ATO12_21745 PE=4 SV=1
MM1 pKa = 7.3ARR3 pKa = 11.84IAGVDD8 pKa = 3.06IPKK11 pKa = 10.08QKK13 pKa = 10.42RR14 pKa = 11.84GVIALTYY21 pKa = 9.53IFGIGRR27 pKa = 11.84SRR29 pKa = 11.84AKK31 pKa = 10.46EE32 pKa = 3.61ILEE35 pKa = 4.06TAKK38 pKa = 10.17VDD40 pKa = 3.69EE41 pKa = 4.55SVKK44 pKa = 10.68VSDD47 pKa = 4.68WDD49 pKa = 3.64DD50 pKa = 3.74DD51 pKa = 4.05QIGRR55 pKa = 11.84IRR57 pKa = 11.84EE58 pKa = 3.97AVGAYY63 pKa = 8.96TIEE66 pKa = 4.71GEE68 pKa = 4.31LRR70 pKa = 11.84SEE72 pKa = 3.94VQLNIKK78 pKa = 10.12RR79 pKa = 11.84LMDD82 pKa = 3.77IGCYY86 pKa = 9.51RR87 pKa = 11.84GIRR90 pKa = 11.84HH91 pKa = 6.49RR92 pKa = 11.84AGLPLRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 11.3NNSRR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.17GRR112 pKa = 11.84RR113 pKa = 11.84KK114 pKa = 7.81TVANKK119 pKa = 10.07KK120 pKa = 9.89KK121 pKa = 9.38ATKK124 pKa = 10.14
MM1 pKa = 7.3ARR3 pKa = 11.84IAGVDD8 pKa = 3.06IPKK11 pKa = 10.08QKK13 pKa = 10.42RR14 pKa = 11.84GVIALTYY21 pKa = 9.53IFGIGRR27 pKa = 11.84SRR29 pKa = 11.84AKK31 pKa = 10.46EE32 pKa = 3.61ILEE35 pKa = 4.06TAKK38 pKa = 10.17VDD40 pKa = 3.69EE41 pKa = 4.55SVKK44 pKa = 10.68VSDD47 pKa = 4.68WDD49 pKa = 3.64DD50 pKa = 3.74DD51 pKa = 4.05QIGRR55 pKa = 11.84IRR57 pKa = 11.84EE58 pKa = 3.97AVGAYY63 pKa = 8.96TIEE66 pKa = 4.71GEE68 pKa = 4.31LRR70 pKa = 11.84SEE72 pKa = 3.94VQLNIKK78 pKa = 10.12RR79 pKa = 11.84LMDD82 pKa = 3.77IGCYY86 pKa = 9.51RR87 pKa = 11.84GIRR90 pKa = 11.84HH91 pKa = 6.49RR92 pKa = 11.84AGLPLRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 11.3NNSRR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.17GRR112 pKa = 11.84RR113 pKa = 11.84KK114 pKa = 7.81TVANKK119 pKa = 10.07KK120 pKa = 9.89KK121 pKa = 9.38ATKK124 pKa = 10.14
Molecular weight: 14.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1642244 |
59 |
7139 |
343.1 |
38.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.875 ± 0.036 | 0.77 ± 0.013 |
5.587 ± 0.026 | 6.526 ± 0.041 |
5.154 ± 0.031 | 6.266 ± 0.04 |
1.828 ± 0.018 | 8.533 ± 0.036 |
7.976 ± 0.048 | 9.142 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.024 ± 0.015 | 6.178 ± 0.042 |
3.314 ± 0.023 | 3.41 ± 0.023 |
3.399 ± 0.023 | 6.656 ± 0.031 |
6.06 ± 0.044 | 5.955 ± 0.026 |
1.068 ± 0.013 | 4.281 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
