
Hubei sobemo-like virus 13
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
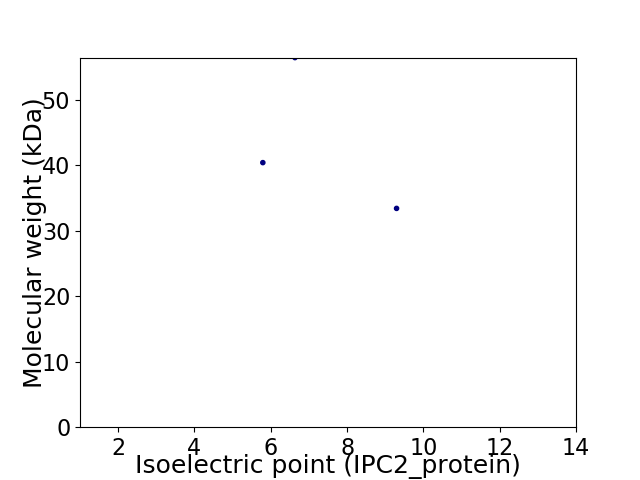
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KF10|A0A1L3KF10_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 13 OX=1923198 PE=4 SV=1
MM1 pKa = 7.35NSVPGVCSLSRR12 pKa = 11.84LGTTNAQVFGMDD24 pKa = 3.13AEE26 pKa = 4.64GRR28 pKa = 11.84FDD30 pKa = 4.51KK31 pKa = 11.21SRR33 pKa = 11.84LDD35 pKa = 3.65LVYY38 pKa = 9.8TAVKK42 pKa = 10.29DD43 pKa = 3.63RR44 pKa = 11.84YY45 pKa = 10.39LAVTQGKK52 pKa = 7.38TEE54 pKa = 4.16SDD56 pKa = 3.5PLKK59 pKa = 10.9VFVKK63 pKa = 9.96QEE65 pKa = 3.67PHH67 pKa = 6.33KK68 pKa = 11.0VEE70 pKa = 3.92KK71 pKa = 10.99LKK73 pKa = 10.77DD74 pKa = 3.37DD75 pKa = 3.89RR76 pKa = 11.84LRR78 pKa = 11.84LIMSVSLVDD87 pKa = 3.79ALVDD91 pKa = 4.61RR92 pKa = 11.84ILFMRR97 pKa = 11.84LANQVIKK104 pKa = 10.79KK105 pKa = 8.14HH106 pKa = 5.15TLTNILIGWSPLGGGYY122 pKa = 10.49RR123 pKa = 11.84LIDD126 pKa = 3.4ALFPGEE132 pKa = 4.62TISIDD137 pKa = 3.14KK138 pKa = 10.46KK139 pKa = 9.49AWDD142 pKa = 3.34WSVPYY147 pKa = 9.7WLLEE151 pKa = 3.89AVKK154 pKa = 10.61GVIKK158 pKa = 10.58RR159 pKa = 11.84LAVDD163 pKa = 4.45APQWWHH169 pKa = 6.83DD170 pKa = 3.65AVDD173 pKa = 3.68TRR175 pKa = 11.84FSCLFGDD182 pKa = 4.17PEE184 pKa = 4.51FVFPDD189 pKa = 3.87GTRR192 pKa = 11.84GRR194 pKa = 11.84QEE196 pKa = 3.48QPGIMKK202 pKa = 10.01SGCYY206 pKa = 8.45LTILINSIAQLVLHH220 pKa = 6.47EE221 pKa = 4.2MAKK224 pKa = 10.38VSLGLGDD231 pKa = 3.67EE232 pKa = 4.4VEE234 pKa = 4.36PVVVLGDD241 pKa = 4.35DD242 pKa = 3.46SLQRR246 pKa = 11.84KK247 pKa = 8.11FDD249 pKa = 3.19RR250 pKa = 11.84WQEE253 pKa = 3.82YY254 pKa = 9.75VDD256 pKa = 4.4YY257 pKa = 9.43ITALGFRR264 pKa = 11.84CEE266 pKa = 4.28VEE268 pKa = 4.03HH269 pKa = 7.56HH270 pKa = 6.35SDD272 pKa = 3.4EE273 pKa = 4.68PEE275 pKa = 3.9FAGFRR280 pKa = 11.84YY281 pKa = 9.52RR282 pKa = 11.84DD283 pKa = 3.61KK284 pKa = 11.09FKK286 pKa = 10.09PAYY289 pKa = 8.58FQKK292 pKa = 10.78HH293 pKa = 5.62LFNLAHH299 pKa = 6.69LTTDD303 pKa = 3.91RR304 pKa = 11.84EE305 pKa = 4.56VAQQTLQNYY314 pKa = 8.23QLLYY318 pKa = 11.1YY319 pKa = 9.92FDD321 pKa = 3.91EE322 pKa = 4.41PMRR325 pKa = 11.84RR326 pKa = 11.84VITDD330 pKa = 3.53MIRR333 pKa = 11.84ILNMPEE339 pKa = 3.36AAADD343 pKa = 3.92EE344 pKa = 4.29LRR346 pKa = 11.84LRR348 pKa = 11.84ALAMGG353 pKa = 4.45
MM1 pKa = 7.35NSVPGVCSLSRR12 pKa = 11.84LGTTNAQVFGMDD24 pKa = 3.13AEE26 pKa = 4.64GRR28 pKa = 11.84FDD30 pKa = 4.51KK31 pKa = 11.21SRR33 pKa = 11.84LDD35 pKa = 3.65LVYY38 pKa = 9.8TAVKK42 pKa = 10.29DD43 pKa = 3.63RR44 pKa = 11.84YY45 pKa = 10.39LAVTQGKK52 pKa = 7.38TEE54 pKa = 4.16SDD56 pKa = 3.5PLKK59 pKa = 10.9VFVKK63 pKa = 9.96QEE65 pKa = 3.67PHH67 pKa = 6.33KK68 pKa = 11.0VEE70 pKa = 3.92KK71 pKa = 10.99LKK73 pKa = 10.77DD74 pKa = 3.37DD75 pKa = 3.89RR76 pKa = 11.84LRR78 pKa = 11.84LIMSVSLVDD87 pKa = 3.79ALVDD91 pKa = 4.61RR92 pKa = 11.84ILFMRR97 pKa = 11.84LANQVIKK104 pKa = 10.79KK105 pKa = 8.14HH106 pKa = 5.15TLTNILIGWSPLGGGYY122 pKa = 10.49RR123 pKa = 11.84LIDD126 pKa = 3.4ALFPGEE132 pKa = 4.62TISIDD137 pKa = 3.14KK138 pKa = 10.46KK139 pKa = 9.49AWDD142 pKa = 3.34WSVPYY147 pKa = 9.7WLLEE151 pKa = 3.89AVKK154 pKa = 10.61GVIKK158 pKa = 10.58RR159 pKa = 11.84LAVDD163 pKa = 4.45APQWWHH169 pKa = 6.83DD170 pKa = 3.65AVDD173 pKa = 3.68TRR175 pKa = 11.84FSCLFGDD182 pKa = 4.17PEE184 pKa = 4.51FVFPDD189 pKa = 3.87GTRR192 pKa = 11.84GRR194 pKa = 11.84QEE196 pKa = 3.48QPGIMKK202 pKa = 10.01SGCYY206 pKa = 8.45LTILINSIAQLVLHH220 pKa = 6.47EE221 pKa = 4.2MAKK224 pKa = 10.38VSLGLGDD231 pKa = 3.67EE232 pKa = 4.4VEE234 pKa = 4.36PVVVLGDD241 pKa = 4.35DD242 pKa = 3.46SLQRR246 pKa = 11.84KK247 pKa = 8.11FDD249 pKa = 3.19RR250 pKa = 11.84WQEE253 pKa = 3.82YY254 pKa = 9.75VDD256 pKa = 4.4YY257 pKa = 9.43ITALGFRR264 pKa = 11.84CEE266 pKa = 4.28VEE268 pKa = 4.03HH269 pKa = 7.56HH270 pKa = 6.35SDD272 pKa = 3.4EE273 pKa = 4.68PEE275 pKa = 3.9FAGFRR280 pKa = 11.84YY281 pKa = 9.52RR282 pKa = 11.84DD283 pKa = 3.61KK284 pKa = 11.09FKK286 pKa = 10.09PAYY289 pKa = 8.58FQKK292 pKa = 10.78HH293 pKa = 5.62LFNLAHH299 pKa = 6.69LTTDD303 pKa = 3.91RR304 pKa = 11.84EE305 pKa = 4.56VAQQTLQNYY314 pKa = 8.23QLLYY318 pKa = 11.1YY319 pKa = 9.92FDD321 pKa = 3.91EE322 pKa = 4.41PMRR325 pKa = 11.84RR326 pKa = 11.84VITDD330 pKa = 3.53MIRR333 pKa = 11.84ILNMPEE339 pKa = 3.36AAADD343 pKa = 3.92EE344 pKa = 4.29LRR346 pKa = 11.84LRR348 pKa = 11.84ALAMGG353 pKa = 4.45
Molecular weight: 40.42 kDa
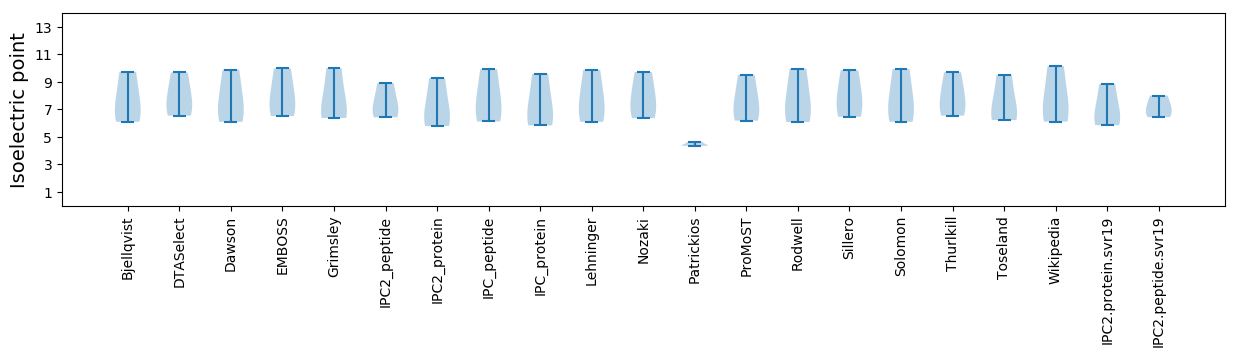
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF19|A0A1L3KF19_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 13 OX=1923198 PE=4 SV=1
MM1 pKa = 7.27LVVLRR6 pKa = 11.84YY7 pKa = 9.61IYY9 pKa = 10.19QKK11 pKa = 11.44GMLKK15 pKa = 9.61VASMLQKK22 pKa = 10.41SSRR25 pKa = 11.84NSKK28 pKa = 9.99LVYY31 pKa = 10.32GSIEE35 pKa = 3.79RR36 pKa = 11.84KK37 pKa = 9.31IRR39 pKa = 11.84IGPSLGMVGAWEE51 pKa = 4.11RR52 pKa = 11.84LSLQLPIILLKK63 pKa = 7.6MTHH66 pKa = 5.62YY67 pKa = 10.84VFAVLRR73 pKa = 11.84AFWTWNLSGFVIWVVTYY90 pKa = 10.14LQLISLTEE98 pKa = 3.91SGLSYY103 pKa = 10.6RR104 pKa = 11.84SRR106 pKa = 11.84LPGFQSWLSLILRR119 pKa = 11.84LYY121 pKa = 8.53MWAVQEE127 pKa = 4.01WHH129 pKa = 6.78LVACLSHH136 pKa = 6.49TLLILTLFMRR146 pKa = 11.84DD147 pKa = 3.2PLNPVFQAHH156 pKa = 6.86LCIQVILYY164 pKa = 7.16MACMWVQALAIWPWILIGSLPLLFATRR191 pKa = 11.84VPRR194 pKa = 11.84SLFSRR199 pKa = 11.84SFLLDD204 pKa = 3.17TNVQEE209 pKa = 4.25RR210 pKa = 11.84TLNSMVLHH218 pKa = 6.16LVNLGSCIMANIAFLMTRR236 pKa = 11.84IYY238 pKa = 10.71QNTYY242 pKa = 8.27GMCWNMLLRR251 pKa = 11.84QLNIMMRR258 pKa = 11.84GSRR261 pKa = 11.84TTHH264 pKa = 6.09FMIDD268 pKa = 3.44QEE270 pKa = 5.86DD271 pKa = 4.53PDD273 pKa = 4.03TDD275 pKa = 3.15QDD277 pKa = 3.93LRR279 pKa = 11.84LMIQPVSEE287 pKa = 4.11
MM1 pKa = 7.27LVVLRR6 pKa = 11.84YY7 pKa = 9.61IYY9 pKa = 10.19QKK11 pKa = 11.44GMLKK15 pKa = 9.61VASMLQKK22 pKa = 10.41SSRR25 pKa = 11.84NSKK28 pKa = 9.99LVYY31 pKa = 10.32GSIEE35 pKa = 3.79RR36 pKa = 11.84KK37 pKa = 9.31IRR39 pKa = 11.84IGPSLGMVGAWEE51 pKa = 4.11RR52 pKa = 11.84LSLQLPIILLKK63 pKa = 7.6MTHH66 pKa = 5.62YY67 pKa = 10.84VFAVLRR73 pKa = 11.84AFWTWNLSGFVIWVVTYY90 pKa = 10.14LQLISLTEE98 pKa = 3.91SGLSYY103 pKa = 10.6RR104 pKa = 11.84SRR106 pKa = 11.84LPGFQSWLSLILRR119 pKa = 11.84LYY121 pKa = 8.53MWAVQEE127 pKa = 4.01WHH129 pKa = 6.78LVACLSHH136 pKa = 6.49TLLILTLFMRR146 pKa = 11.84DD147 pKa = 3.2PLNPVFQAHH156 pKa = 6.86LCIQVILYY164 pKa = 7.16MACMWVQALAIWPWILIGSLPLLFATRR191 pKa = 11.84VPRR194 pKa = 11.84SLFSRR199 pKa = 11.84SFLLDD204 pKa = 3.17TNVQEE209 pKa = 4.25RR210 pKa = 11.84TLNSMVLHH218 pKa = 6.16LVNLGSCIMANIAFLMTRR236 pKa = 11.84IYY238 pKa = 10.71QNTYY242 pKa = 8.27GMCWNMLLRR251 pKa = 11.84QLNIMMRR258 pKa = 11.84GSRR261 pKa = 11.84TTHH264 pKa = 6.09FMIDD268 pKa = 3.44QEE270 pKa = 5.86DD271 pKa = 4.53PDD273 pKa = 4.03TDD275 pKa = 3.15QDD277 pKa = 3.93LRR279 pKa = 11.84LMIQPVSEE287 pKa = 4.11
Molecular weight: 33.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1144 |
287 |
504 |
381.3 |
43.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.381 ± 0.606 | 1.311 ± 0.14 |
5.07 ± 1.244 | 5.157 ± 0.899 |
4.371 ± 0.212 | 6.381 ± 0.821 |
2.273 ± 0.067 | 6.731 ± 0.812 |
4.196 ± 0.765 | 10.664 ± 2.52 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.234 ± 1.024 | 2.972 ± 0.296 |
4.808 ± 0.63 | 4.633 ± 0.21 |
6.643 ± 0.068 | 6.906 ± 0.986 |
5.157 ± 0.343 | 7.168 ± 0.455 |
2.535 ± 0.423 | 3.409 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
