
Burkholderia phage phiE52237
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Peduovirinae; Tigrvirus; Burkholderia virus phi52237
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
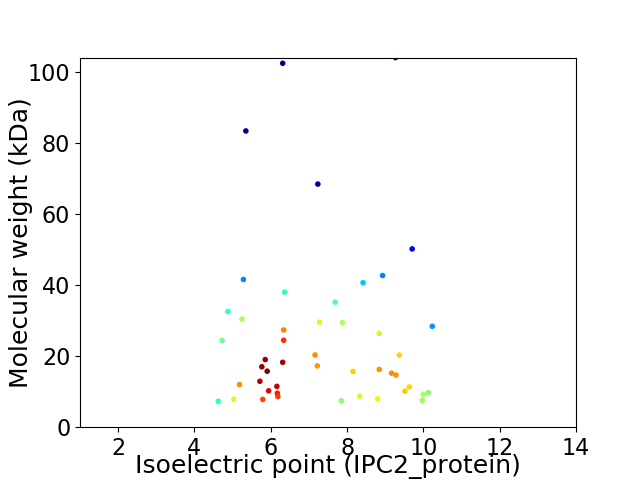
Virtual 2D-PAGE plot for 47 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q45YF1|Q45YF1_9CAUD Putative bacteriophage membrane protein OS=Burkholderia phage phiE52237 OX=2681592 GN=BPSphi5223_0037 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 10.02IAALQGEE9 pKa = 4.7TLDD12 pKa = 3.91ALCWRR17 pKa = 11.84HH18 pKa = 5.61YY19 pKa = 10.73GSTAGTVEE27 pKa = 4.42AVLEE31 pKa = 4.34ANPGLAEE38 pKa = 4.5LGVVLPMGTVVEE50 pKa = 4.43MPEE53 pKa = 3.54RR54 pKa = 11.84RR55 pKa = 11.84AIEE58 pKa = 4.11TTTPLLQLFDD68 pKa = 3.82
MM1 pKa = 7.69KK2 pKa = 10.02IAALQGEE9 pKa = 4.7TLDD12 pKa = 3.91ALCWRR17 pKa = 11.84HH18 pKa = 5.61YY19 pKa = 10.73GSTAGTVEE27 pKa = 4.42AVLEE31 pKa = 4.34ANPGLAEE38 pKa = 4.5LGVVLPMGTVVEE50 pKa = 4.43MPEE53 pKa = 3.54RR54 pKa = 11.84RR55 pKa = 11.84AIEE58 pKa = 4.11TTTPLLQLFDD68 pKa = 3.82
Molecular weight: 7.3 kDa
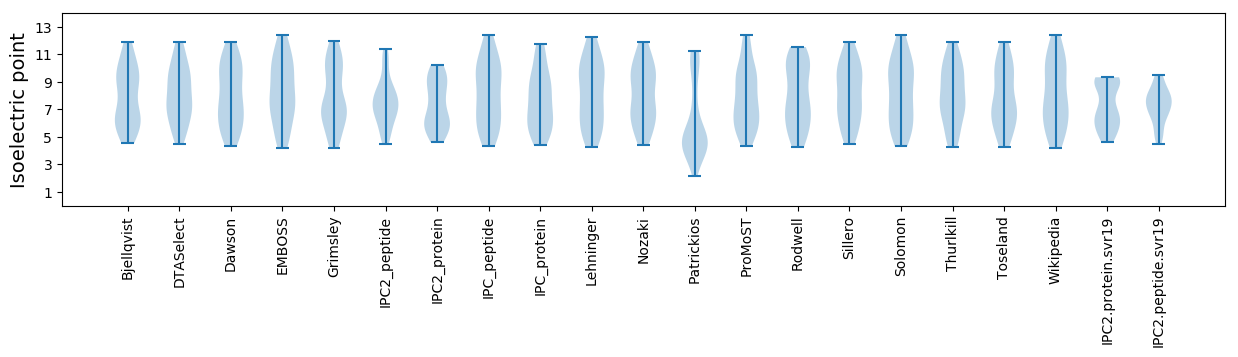
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q45YI7|Q45YI7_9CAUD Uncharacterized protein OS=Burkholderia phage phiE52237 OX=2681592 GN=BPSphi5223_0001 PE=4 SV=1
MM1 pKa = 7.03VVHH4 pKa = 7.21DD5 pKa = 3.84ARR7 pKa = 11.84NRR9 pKa = 11.84VRR11 pKa = 11.84MHH13 pKa = 6.76PPPRR17 pKa = 11.84TAGRR21 pKa = 11.84KK22 pKa = 8.96DD23 pKa = 3.33RR24 pKa = 11.84SSVGSRR30 pKa = 11.84GCRR33 pKa = 11.84RR34 pKa = 11.84DD35 pKa = 3.7RR36 pKa = 11.84LAQTAIRR43 pKa = 11.84KK44 pKa = 8.07NRR46 pKa = 11.84WEE48 pKa = 3.47MRR50 pKa = 11.84FYY52 pKa = 10.93KK53 pKa = 10.08RR54 pKa = 11.84QKK56 pKa = 9.85NGPWWFDD63 pKa = 3.05LSIDD67 pKa = 3.7GSRR70 pKa = 11.84IRR72 pKa = 11.84QSSGTSDD79 pKa = 2.75RR80 pKa = 11.84RR81 pKa = 11.84AAEE84 pKa = 3.71EE85 pKa = 3.76LAAKK89 pKa = 9.58VASDD93 pKa = 2.99YY94 pKa = 10.33WRR96 pKa = 11.84QKK98 pKa = 11.15KK99 pKa = 10.23LGEE102 pKa = 4.24RR103 pKa = 11.84PAVTWDD109 pKa = 3.13AAVVHH114 pKa = 6.13WLKK117 pKa = 10.89QNQHH121 pKa = 4.62QRR123 pKa = 11.84SLEE126 pKa = 4.1TTKK129 pKa = 10.4QRR131 pKa = 11.84LRR133 pKa = 11.84WLTDD137 pKa = 3.02QLKK140 pKa = 10.66GEE142 pKa = 4.22NVRR145 pKa = 11.84NIDD148 pKa = 3.52RR149 pKa = 11.84EE150 pKa = 4.15RR151 pKa = 11.84IQALIEE157 pKa = 3.81TKK159 pKa = 10.45AAEE162 pKa = 4.5KK163 pKa = 10.88YY164 pKa = 10.03RR165 pKa = 11.84GSPVAGATVNRR176 pKa = 11.84HH177 pKa = 4.03MAALSVILHH186 pKa = 6.28HH187 pKa = 6.37CHH189 pKa = 6.46AEE191 pKa = 3.93GWIDD195 pKa = 3.67AVPPIRR201 pKa = 11.84KK202 pKa = 8.83LRR204 pKa = 11.84EE205 pKa = 3.33NSARR209 pKa = 11.84LTWLTRR215 pKa = 11.84AQAQRR220 pKa = 11.84LLEE223 pKa = 4.15EE224 pKa = 4.95LPTHH228 pKa = 6.55LRR230 pKa = 11.84QMARR234 pKa = 11.84FALATGLRR242 pKa = 11.84EE243 pKa = 4.27SNVRR247 pKa = 11.84LLEE250 pKa = 3.9WAQVDD255 pKa = 4.41RR256 pKa = 11.84EE257 pKa = 4.03RR258 pKa = 11.84ALAWIHH264 pKa = 6.96ADD266 pKa = 3.13QAKK269 pKa = 9.28AGKK272 pKa = 9.85VISVPLNEE280 pKa = 4.97DD281 pKa = 2.97ALGVLRR287 pKa = 11.84EE288 pKa = 3.99QQGQHH293 pKa = 4.65KK294 pKa = 10.04RR295 pKa = 11.84YY296 pKa = 8.69VFVYY300 pKa = 9.12KK301 pKa = 10.36GAPIGRR307 pKa = 11.84IYY309 pKa = 10.26NHH311 pKa = 6.83AWQKK315 pKa = 9.5ACVRR319 pKa = 11.84AGLSGLRR326 pKa = 11.84FHH328 pKa = 7.58DD329 pKa = 5.35LRR331 pKa = 11.84HH332 pKa = 5.71TWASWHH338 pKa = 5.46VQAGTPLPILQQLGGWASYY357 pKa = 10.9QMVLRR362 pKa = 11.84YY363 pKa = 9.69AHH365 pKa = 7.51LGRR368 pKa = 11.84DD369 pKa = 3.49HH370 pKa = 6.31VAAYY374 pKa = 10.25ADD376 pKa = 3.74NIGTLRR382 pKa = 11.84HH383 pKa = 6.08KK384 pKa = 10.73SGTPPEE390 pKa = 4.1IEE392 pKa = 4.42KK393 pKa = 11.01GSDD396 pKa = 3.24CSEE399 pKa = 4.09PLSNLVAWGGIEE411 pKa = 3.84PPTRR415 pKa = 11.84GFSKK419 pKa = 9.85RR420 pKa = 11.84TADD423 pKa = 3.31TSAPSGKK430 pKa = 10.14ASRR433 pKa = 11.84PPKK436 pKa = 9.56KK437 pKa = 10.13RR438 pKa = 11.84KK439 pKa = 9.06AAA441 pKa = 3.76
MM1 pKa = 7.03VVHH4 pKa = 7.21DD5 pKa = 3.84ARR7 pKa = 11.84NRR9 pKa = 11.84VRR11 pKa = 11.84MHH13 pKa = 6.76PPPRR17 pKa = 11.84TAGRR21 pKa = 11.84KK22 pKa = 8.96DD23 pKa = 3.33RR24 pKa = 11.84SSVGSRR30 pKa = 11.84GCRR33 pKa = 11.84RR34 pKa = 11.84DD35 pKa = 3.7RR36 pKa = 11.84LAQTAIRR43 pKa = 11.84KK44 pKa = 8.07NRR46 pKa = 11.84WEE48 pKa = 3.47MRR50 pKa = 11.84FYY52 pKa = 10.93KK53 pKa = 10.08RR54 pKa = 11.84QKK56 pKa = 9.85NGPWWFDD63 pKa = 3.05LSIDD67 pKa = 3.7GSRR70 pKa = 11.84IRR72 pKa = 11.84QSSGTSDD79 pKa = 2.75RR80 pKa = 11.84RR81 pKa = 11.84AAEE84 pKa = 3.71EE85 pKa = 3.76LAAKK89 pKa = 9.58VASDD93 pKa = 2.99YY94 pKa = 10.33WRR96 pKa = 11.84QKK98 pKa = 11.15KK99 pKa = 10.23LGEE102 pKa = 4.24RR103 pKa = 11.84PAVTWDD109 pKa = 3.13AAVVHH114 pKa = 6.13WLKK117 pKa = 10.89QNQHH121 pKa = 4.62QRR123 pKa = 11.84SLEE126 pKa = 4.1TTKK129 pKa = 10.4QRR131 pKa = 11.84LRR133 pKa = 11.84WLTDD137 pKa = 3.02QLKK140 pKa = 10.66GEE142 pKa = 4.22NVRR145 pKa = 11.84NIDD148 pKa = 3.52RR149 pKa = 11.84EE150 pKa = 4.15RR151 pKa = 11.84IQALIEE157 pKa = 3.81TKK159 pKa = 10.45AAEE162 pKa = 4.5KK163 pKa = 10.88YY164 pKa = 10.03RR165 pKa = 11.84GSPVAGATVNRR176 pKa = 11.84HH177 pKa = 4.03MAALSVILHH186 pKa = 6.28HH187 pKa = 6.37CHH189 pKa = 6.46AEE191 pKa = 3.93GWIDD195 pKa = 3.67AVPPIRR201 pKa = 11.84KK202 pKa = 8.83LRR204 pKa = 11.84EE205 pKa = 3.33NSARR209 pKa = 11.84LTWLTRR215 pKa = 11.84AQAQRR220 pKa = 11.84LLEE223 pKa = 4.15EE224 pKa = 4.95LPTHH228 pKa = 6.55LRR230 pKa = 11.84QMARR234 pKa = 11.84FALATGLRR242 pKa = 11.84EE243 pKa = 4.27SNVRR247 pKa = 11.84LLEE250 pKa = 3.9WAQVDD255 pKa = 4.41RR256 pKa = 11.84EE257 pKa = 4.03RR258 pKa = 11.84ALAWIHH264 pKa = 6.96ADD266 pKa = 3.13QAKK269 pKa = 9.28AGKK272 pKa = 9.85VISVPLNEE280 pKa = 4.97DD281 pKa = 2.97ALGVLRR287 pKa = 11.84EE288 pKa = 3.99QQGQHH293 pKa = 4.65KK294 pKa = 10.04RR295 pKa = 11.84YY296 pKa = 8.69VFVYY300 pKa = 9.12KK301 pKa = 10.36GAPIGRR307 pKa = 11.84IYY309 pKa = 10.26NHH311 pKa = 6.83AWQKK315 pKa = 9.5ACVRR319 pKa = 11.84AGLSGLRR326 pKa = 11.84FHH328 pKa = 7.58DD329 pKa = 5.35LRR331 pKa = 11.84HH332 pKa = 5.71TWASWHH338 pKa = 5.46VQAGTPLPILQQLGGWASYY357 pKa = 10.9QMVLRR362 pKa = 11.84YY363 pKa = 9.69AHH365 pKa = 7.51LGRR368 pKa = 11.84DD369 pKa = 3.49HH370 pKa = 6.31VAAYY374 pKa = 10.25ADD376 pKa = 3.74NIGTLRR382 pKa = 11.84HH383 pKa = 6.08KK384 pKa = 10.73SGTPPEE390 pKa = 4.1IEE392 pKa = 4.42KK393 pKa = 11.01GSDD396 pKa = 3.24CSEE399 pKa = 4.09PLSNLVAWGGIEE411 pKa = 3.84PPTRR415 pKa = 11.84GFSKK419 pKa = 9.85RR420 pKa = 11.84TADD423 pKa = 3.31TSAPSGKK430 pKa = 10.14ASRR433 pKa = 11.84PPKK436 pKa = 9.56KK437 pKa = 10.13RR438 pKa = 11.84KK439 pKa = 9.06AAA441 pKa = 3.76
Molecular weight: 50.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11038 |
65 |
988 |
234.9 |
25.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.526 ± 0.526 | 0.843 ± 0.134 |
6.161 ± 0.385 | 5.2 ± 0.265 |
3.28 ± 0.254 | 7.9 ± 0.525 |
2.482 ± 0.204 | 3.95 ± 0.221 |
4.05 ± 0.311 | 8.942 ± 0.389 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.355 ± 0.187 | 2.935 ± 0.182 |
4.765 ± 0.373 | 3.434 ± 0.267 |
8.462 ± 0.507 | 5.064 ± 0.226 |
5.862 ± 0.305 | 6.777 ± 0.283 |
1.54 ± 0.171 | 2.473 ± 0.168 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
