
WU polyomavirus (WUPyV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus; Human polyomavirus 4
Average proteome isoelectric point is 7.56
Get precalculated fractions of proteins
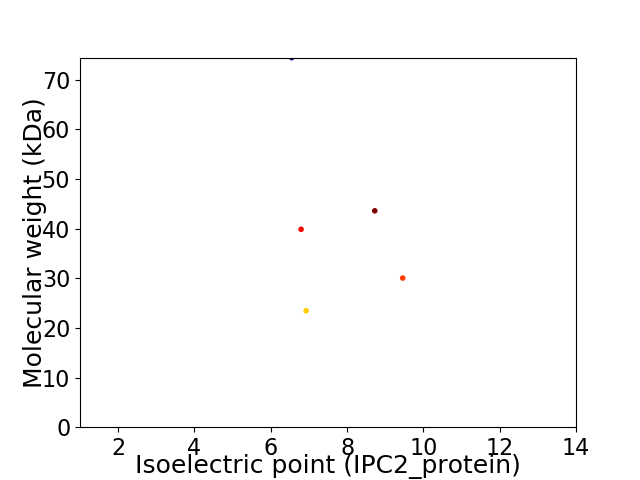
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
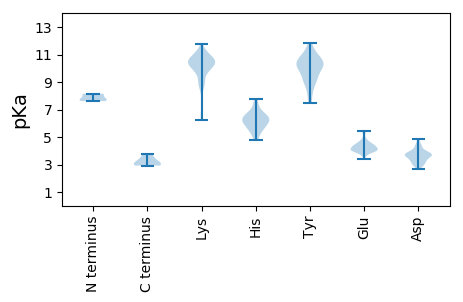
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|A5HBE1-2|VP2-2_POVWU Isoform of A5HBE1 Isoform VP3 of Minor capsid protein VP2 OS=WU polyomavirus OX=440266 PE=3 SV=1
MM1 pKa = 8.15DD2 pKa = 3.67KK3 pKa = 10.13TLSRR7 pKa = 11.84NEE9 pKa = 4.16AKK11 pKa = 10.6EE12 pKa = 3.79LMQLLGLDD20 pKa = 3.57MTCWGNLPLMRR31 pKa = 11.84TKK33 pKa = 10.78YY34 pKa = 10.37LSKK37 pKa = 10.64CKK39 pKa = 9.91EE40 pKa = 3.7FHH42 pKa = 6.72PDD44 pKa = 2.82KK45 pKa = 11.16GGNEE49 pKa = 3.91EE50 pKa = 4.92KK51 pKa = 10.12MKK53 pKa = 10.72KK54 pKa = 10.31LNSLYY59 pKa = 10.78LKK61 pKa = 10.45LQEE64 pKa = 4.67CVSTVHH70 pKa = 6.14QLNEE74 pKa = 4.13EE75 pKa = 3.99EE76 pKa = 4.62DD77 pKa = 4.45EE78 pKa = 4.47VWSSSQIPTYY88 pKa = 8.7GTPDD92 pKa = 3.01WDD94 pKa = 4.04YY95 pKa = 10.23WWSQFNSYY103 pKa = 8.75WEE105 pKa = 4.03EE106 pKa = 3.58EE107 pKa = 4.2LRR109 pKa = 11.84CNEE112 pKa = 4.56EE113 pKa = 4.12MPKK116 pKa = 10.4SPGEE120 pKa = 4.27TPTKK124 pKa = 9.6RR125 pKa = 11.84TRR127 pKa = 11.84EE128 pKa = 3.95DD129 pKa = 3.69DD130 pKa = 4.17EE131 pKa = 5.35EE132 pKa = 4.44PQCSQATPPKK142 pKa = 9.75KK143 pKa = 10.5KK144 pKa = 9.99KK145 pKa = 10.64DD146 pKa = 3.66NATDD150 pKa = 3.52ASLSFPKK157 pKa = 10.28EE158 pKa = 3.61LEE160 pKa = 4.15EE161 pKa = 4.38FVSQAVFSNRR171 pKa = 11.84TLTAFVIHH179 pKa = 5.2TTKK182 pKa = 10.73EE183 pKa = 4.0KK184 pKa = 11.34AEE186 pKa = 4.03TLYY189 pKa = 11.18KK190 pKa = 10.61KK191 pKa = 10.53LLSKK195 pKa = 8.86FKK197 pKa = 11.03CNFASRR203 pKa = 11.84HH204 pKa = 4.86SYY206 pKa = 11.01YY207 pKa = 9.85NTALVFILTPFRR219 pKa = 11.84HH220 pKa = 5.52RR221 pKa = 11.84VSAVNNFCKK230 pKa = 10.39GYY232 pKa = 8.56CTISFLFCKK241 pKa = 10.24GVNNAYY247 pKa = 10.36GLYY250 pKa = 10.57SRR252 pKa = 11.84MTRR255 pKa = 11.84DD256 pKa = 3.26PFTLCEE262 pKa = 3.9EE263 pKa = 4.69NIPGGLKK270 pKa = 10.74EE271 pKa = 4.42NDD273 pKa = 3.48FKK275 pKa = 11.76AEE277 pKa = 3.95DD278 pKa = 3.92LYY280 pKa = 11.77GEE282 pKa = 5.23FKK284 pKa = 11.04DD285 pKa = 3.73QLNWKK290 pKa = 9.1ALSEE294 pKa = 4.19FALEE298 pKa = 4.4LGIDD302 pKa = 3.86DD303 pKa = 4.83VYY305 pKa = 11.82LLLGLYY311 pKa = 9.53LQLSIKK317 pKa = 10.48VEE319 pKa = 4.04EE320 pKa = 4.47CEE322 pKa = 4.1KK323 pKa = 11.13CNSNEE328 pKa = 4.03DD329 pKa = 3.37ATHH332 pKa = 6.02NRR334 pKa = 11.84LHH336 pKa = 6.52MEE338 pKa = 3.95HH339 pKa = 6.28QKK341 pKa = 10.75NALLFSDD348 pKa = 4.63SKK350 pKa = 10.58SQKK353 pKa = 9.84NVCQQAIDD361 pKa = 3.6VVIAKK366 pKa = 10.07RR367 pKa = 11.84RR368 pKa = 11.84VDD370 pKa = 3.72SLNMSRR376 pKa = 11.84EE377 pKa = 4.02DD378 pKa = 3.25LLARR382 pKa = 11.84RR383 pKa = 11.84FEE385 pKa = 4.73KK386 pKa = 10.62ILDD389 pKa = 3.97KK390 pKa = 10.18MDD392 pKa = 3.31KK393 pKa = 9.68TIKK396 pKa = 10.72GEE398 pKa = 4.05QDD400 pKa = 3.12VLLYY404 pKa = 9.12MAGVAWYY411 pKa = 9.99LGLNGKK417 pKa = 9.09IDD419 pKa = 3.84EE420 pKa = 4.33LVYY423 pKa = 10.41RR424 pKa = 11.84YY425 pKa = 10.56LKK427 pKa = 10.76VIVEE431 pKa = 4.24NVPKK435 pKa = 10.22KK436 pKa = 10.46RR437 pKa = 11.84YY438 pKa = 8.27WVFKK442 pKa = 10.85GPINSGKK449 pKa = 6.84TTVAAALLDD458 pKa = 3.9LCGGKK463 pKa = 9.81ALNINIPADD472 pKa = 3.71RR473 pKa = 11.84LNFEE477 pKa = 5.01LGVAIDD483 pKa = 3.39QFTVVFEE490 pKa = 4.28DD491 pKa = 3.34VKK493 pKa = 11.44GQVGDD498 pKa = 3.88NKK500 pKa = 10.6LLPSGNGMSNLDD512 pKa = 3.72NLRR515 pKa = 11.84DD516 pKa = 3.83YY517 pKa = 11.6LDD519 pKa = 4.13GSVKK523 pKa = 10.37VNLEE527 pKa = 3.88KK528 pKa = 10.8KK529 pKa = 9.64HH530 pKa = 5.73LNKK533 pKa = 10.08RR534 pKa = 11.84SQIFPPGIVTMNEE547 pKa = 3.84YY548 pKa = 10.54LVPATLAPRR557 pKa = 11.84FHH559 pKa = 6.4KK560 pKa = 9.6TVLFTPKK567 pKa = 9.88RR568 pKa = 11.84HH569 pKa = 6.07LKK571 pKa = 10.32EE572 pKa = 4.64SLDD575 pKa = 3.63KK576 pKa = 11.12TPEE579 pKa = 3.96LMVKK583 pKa = 10.31RR584 pKa = 11.84VLQSGMCILLMLIWCRR600 pKa = 11.84PVSDD604 pKa = 4.39FHH606 pKa = 7.77PCIQAKK612 pKa = 8.84VVYY615 pKa = 9.2WKK617 pKa = 10.62EE618 pKa = 3.94LLDD621 pKa = 3.9KK622 pKa = 11.22YY623 pKa = 10.92IGLTEE628 pKa = 4.07FADD631 pKa = 3.43MQMNVTNGCNILEE644 pKa = 4.21KK645 pKa = 10.92HH646 pKa = 5.58NAA648 pKa = 3.31
MM1 pKa = 8.15DD2 pKa = 3.67KK3 pKa = 10.13TLSRR7 pKa = 11.84NEE9 pKa = 4.16AKK11 pKa = 10.6EE12 pKa = 3.79LMQLLGLDD20 pKa = 3.57MTCWGNLPLMRR31 pKa = 11.84TKK33 pKa = 10.78YY34 pKa = 10.37LSKK37 pKa = 10.64CKK39 pKa = 9.91EE40 pKa = 3.7FHH42 pKa = 6.72PDD44 pKa = 2.82KK45 pKa = 11.16GGNEE49 pKa = 3.91EE50 pKa = 4.92KK51 pKa = 10.12MKK53 pKa = 10.72KK54 pKa = 10.31LNSLYY59 pKa = 10.78LKK61 pKa = 10.45LQEE64 pKa = 4.67CVSTVHH70 pKa = 6.14QLNEE74 pKa = 4.13EE75 pKa = 3.99EE76 pKa = 4.62DD77 pKa = 4.45EE78 pKa = 4.47VWSSSQIPTYY88 pKa = 8.7GTPDD92 pKa = 3.01WDD94 pKa = 4.04YY95 pKa = 10.23WWSQFNSYY103 pKa = 8.75WEE105 pKa = 4.03EE106 pKa = 3.58EE107 pKa = 4.2LRR109 pKa = 11.84CNEE112 pKa = 4.56EE113 pKa = 4.12MPKK116 pKa = 10.4SPGEE120 pKa = 4.27TPTKK124 pKa = 9.6RR125 pKa = 11.84TRR127 pKa = 11.84EE128 pKa = 3.95DD129 pKa = 3.69DD130 pKa = 4.17EE131 pKa = 5.35EE132 pKa = 4.44PQCSQATPPKK142 pKa = 9.75KK143 pKa = 10.5KK144 pKa = 9.99KK145 pKa = 10.64DD146 pKa = 3.66NATDD150 pKa = 3.52ASLSFPKK157 pKa = 10.28EE158 pKa = 3.61LEE160 pKa = 4.15EE161 pKa = 4.38FVSQAVFSNRR171 pKa = 11.84TLTAFVIHH179 pKa = 5.2TTKK182 pKa = 10.73EE183 pKa = 4.0KK184 pKa = 11.34AEE186 pKa = 4.03TLYY189 pKa = 11.18KK190 pKa = 10.61KK191 pKa = 10.53LLSKK195 pKa = 8.86FKK197 pKa = 11.03CNFASRR203 pKa = 11.84HH204 pKa = 4.86SYY206 pKa = 11.01YY207 pKa = 9.85NTALVFILTPFRR219 pKa = 11.84HH220 pKa = 5.52RR221 pKa = 11.84VSAVNNFCKK230 pKa = 10.39GYY232 pKa = 8.56CTISFLFCKK241 pKa = 10.24GVNNAYY247 pKa = 10.36GLYY250 pKa = 10.57SRR252 pKa = 11.84MTRR255 pKa = 11.84DD256 pKa = 3.26PFTLCEE262 pKa = 3.9EE263 pKa = 4.69NIPGGLKK270 pKa = 10.74EE271 pKa = 4.42NDD273 pKa = 3.48FKK275 pKa = 11.76AEE277 pKa = 3.95DD278 pKa = 3.92LYY280 pKa = 11.77GEE282 pKa = 5.23FKK284 pKa = 11.04DD285 pKa = 3.73QLNWKK290 pKa = 9.1ALSEE294 pKa = 4.19FALEE298 pKa = 4.4LGIDD302 pKa = 3.86DD303 pKa = 4.83VYY305 pKa = 11.82LLLGLYY311 pKa = 9.53LQLSIKK317 pKa = 10.48VEE319 pKa = 4.04EE320 pKa = 4.47CEE322 pKa = 4.1KK323 pKa = 11.13CNSNEE328 pKa = 4.03DD329 pKa = 3.37ATHH332 pKa = 6.02NRR334 pKa = 11.84LHH336 pKa = 6.52MEE338 pKa = 3.95HH339 pKa = 6.28QKK341 pKa = 10.75NALLFSDD348 pKa = 4.63SKK350 pKa = 10.58SQKK353 pKa = 9.84NVCQQAIDD361 pKa = 3.6VVIAKK366 pKa = 10.07RR367 pKa = 11.84RR368 pKa = 11.84VDD370 pKa = 3.72SLNMSRR376 pKa = 11.84EE377 pKa = 4.02DD378 pKa = 3.25LLARR382 pKa = 11.84RR383 pKa = 11.84FEE385 pKa = 4.73KK386 pKa = 10.62ILDD389 pKa = 3.97KK390 pKa = 10.18MDD392 pKa = 3.31KK393 pKa = 9.68TIKK396 pKa = 10.72GEE398 pKa = 4.05QDD400 pKa = 3.12VLLYY404 pKa = 9.12MAGVAWYY411 pKa = 9.99LGLNGKK417 pKa = 9.09IDD419 pKa = 3.84EE420 pKa = 4.33LVYY423 pKa = 10.41RR424 pKa = 11.84YY425 pKa = 10.56LKK427 pKa = 10.76VIVEE431 pKa = 4.24NVPKK435 pKa = 10.22KK436 pKa = 10.46RR437 pKa = 11.84YY438 pKa = 8.27WVFKK442 pKa = 10.85GPINSGKK449 pKa = 6.84TTVAAALLDD458 pKa = 3.9LCGGKK463 pKa = 9.81ALNINIPADD472 pKa = 3.71RR473 pKa = 11.84LNFEE477 pKa = 5.01LGVAIDD483 pKa = 3.39QFTVVFEE490 pKa = 4.28DD491 pKa = 3.34VKK493 pKa = 11.44GQVGDD498 pKa = 3.88NKK500 pKa = 10.6LLPSGNGMSNLDD512 pKa = 3.72NLRR515 pKa = 11.84DD516 pKa = 3.83YY517 pKa = 11.6LDD519 pKa = 4.13GSVKK523 pKa = 10.37VNLEE527 pKa = 3.88KK528 pKa = 10.8KK529 pKa = 9.64HH530 pKa = 5.73LNKK533 pKa = 10.08RR534 pKa = 11.84SQIFPPGIVTMNEE547 pKa = 3.84YY548 pKa = 10.54LVPATLAPRR557 pKa = 11.84FHH559 pKa = 6.4KK560 pKa = 9.6TVLFTPKK567 pKa = 9.88RR568 pKa = 11.84HH569 pKa = 6.07LKK571 pKa = 10.32EE572 pKa = 4.64SLDD575 pKa = 3.63KK576 pKa = 11.12TPEE579 pKa = 3.96LMVKK583 pKa = 10.31RR584 pKa = 11.84VLQSGMCILLMLIWCRR600 pKa = 11.84PVSDD604 pKa = 4.39FHH606 pKa = 7.77PCIQAKK612 pKa = 8.84VVYY615 pKa = 9.2WKK617 pKa = 10.62EE618 pKa = 3.94LLDD621 pKa = 3.9KK622 pKa = 11.22YY623 pKa = 10.92IGLTEE628 pKa = 4.07FADD631 pKa = 3.43MQMNVTNGCNILEE644 pKa = 4.21KK645 pKa = 10.92HH646 pKa = 5.58NAA648 pKa = 3.31
Molecular weight: 74.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A5HBE1-2|VP2-2_POVWU Isoform of A5HBE1 Isoform VP3 of Minor capsid protein VP2 OS=WU polyomavirus OX=440266 PE=3 SV=1
MM1 pKa = 7.61ALAPIPEE8 pKa = 4.39YY9 pKa = 10.8NLEE12 pKa = 4.07TGIPGVPDD20 pKa = 3.05WVFNFIASHH29 pKa = 5.87LPEE32 pKa = 5.33LPSLQDD38 pKa = 3.05VFNRR42 pKa = 11.84IAYY45 pKa = 7.47GIWTSYY51 pKa = 10.35YY52 pKa = 7.9NTGRR56 pKa = 11.84TVVNRR61 pKa = 11.84AVSEE65 pKa = 3.97EE66 pKa = 3.96LQRR69 pKa = 11.84LLGDD73 pKa = 3.59LEE75 pKa = 4.56YY76 pKa = 11.22GFRR79 pKa = 11.84TALATIGEE87 pKa = 4.54SDD89 pKa = 3.83PVNAIVEE96 pKa = 4.38QVRR99 pKa = 11.84SFVSGGRR106 pKa = 11.84QRR108 pKa = 11.84EE109 pKa = 4.13LLQIAAGQPVDD120 pKa = 3.09ISEE123 pKa = 4.26GVSRR127 pKa = 11.84GTATISNAVEE137 pKa = 3.96AVRR140 pKa = 11.84DD141 pKa = 3.67ATQRR145 pKa = 11.84LSQATYY151 pKa = 10.27NFVYY155 pKa = 10.39DD156 pKa = 4.26ASTLPRR162 pKa = 11.84DD163 pKa = 3.84GFNALSDD170 pKa = 3.79GVHH173 pKa = 7.37RR174 pKa = 11.84LGQWISMPGATGGTPHH190 pKa = 6.65YY191 pKa = 9.8AAPDD195 pKa = 3.69WILYY199 pKa = 9.04VLEE202 pKa = 4.79EE203 pKa = 4.48LNSDD207 pKa = 2.93ISKK210 pKa = 10.48IPTQGIKK217 pKa = 10.29RR218 pKa = 11.84KK219 pKa = 8.7LQQNGLHH226 pKa = 6.28SKK228 pKa = 10.93ASLHH232 pKa = 5.72SKK234 pKa = 6.67TRR236 pKa = 11.84KK237 pKa = 6.24VTKK240 pKa = 10.4KK241 pKa = 8.99STHH244 pKa = 6.19KK245 pKa = 10.24SAKK248 pKa = 9.15PSKK251 pKa = 8.8TSQKK255 pKa = 9.88RR256 pKa = 11.84RR257 pKa = 11.84GRR259 pKa = 11.84RR260 pKa = 11.84AGRR263 pKa = 11.84RR264 pKa = 11.84TTVRR268 pKa = 11.84RR269 pKa = 11.84NRR271 pKa = 11.84VV272 pKa = 2.93
MM1 pKa = 7.61ALAPIPEE8 pKa = 4.39YY9 pKa = 10.8NLEE12 pKa = 4.07TGIPGVPDD20 pKa = 3.05WVFNFIASHH29 pKa = 5.87LPEE32 pKa = 5.33LPSLQDD38 pKa = 3.05VFNRR42 pKa = 11.84IAYY45 pKa = 7.47GIWTSYY51 pKa = 10.35YY52 pKa = 7.9NTGRR56 pKa = 11.84TVVNRR61 pKa = 11.84AVSEE65 pKa = 3.97EE66 pKa = 3.96LQRR69 pKa = 11.84LLGDD73 pKa = 3.59LEE75 pKa = 4.56YY76 pKa = 11.22GFRR79 pKa = 11.84TALATIGEE87 pKa = 4.54SDD89 pKa = 3.83PVNAIVEE96 pKa = 4.38QVRR99 pKa = 11.84SFVSGGRR106 pKa = 11.84QRR108 pKa = 11.84EE109 pKa = 4.13LLQIAAGQPVDD120 pKa = 3.09ISEE123 pKa = 4.26GVSRR127 pKa = 11.84GTATISNAVEE137 pKa = 3.96AVRR140 pKa = 11.84DD141 pKa = 3.67ATQRR145 pKa = 11.84LSQATYY151 pKa = 10.27NFVYY155 pKa = 10.39DD156 pKa = 4.26ASTLPRR162 pKa = 11.84DD163 pKa = 3.84GFNALSDD170 pKa = 3.79GVHH173 pKa = 7.37RR174 pKa = 11.84LGQWISMPGATGGTPHH190 pKa = 6.65YY191 pKa = 9.8AAPDD195 pKa = 3.69WILYY199 pKa = 9.04VLEE202 pKa = 4.79EE203 pKa = 4.48LNSDD207 pKa = 2.93ISKK210 pKa = 10.48IPTQGIKK217 pKa = 10.29RR218 pKa = 11.84KK219 pKa = 8.7LQQNGLHH226 pKa = 6.28SKK228 pKa = 10.93ASLHH232 pKa = 5.72SKK234 pKa = 6.67TRR236 pKa = 11.84KK237 pKa = 6.24VTKK240 pKa = 10.4KK241 pKa = 8.99STHH244 pKa = 6.19KK245 pKa = 10.24SAKK248 pKa = 9.15PSKK251 pKa = 8.8TSQKK255 pKa = 9.88RR256 pKa = 11.84RR257 pKa = 11.84GRR259 pKa = 11.84RR260 pKa = 11.84AGRR263 pKa = 11.84RR264 pKa = 11.84TTVRR268 pKa = 11.84RR269 pKa = 11.84NRR271 pKa = 11.84VV272 pKa = 2.93
Molecular weight: 30.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1898 |
194 |
648 |
379.6 |
42.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.902 ± 1.544 | 2.002 ± 0.911 |
4.268 ± 0.572 | 6.428 ± 0.775 |
3.319 ± 0.651 | 7.376 ± 1.193 |
1.897 ± 0.16 | 4.426 ± 0.484 |
6.375 ± 1.414 | 10.063 ± 0.751 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.844 ± 0.506 | 4.953 ± 0.643 |
5.532 ± 0.899 | 3.793 ± 0.276 |
5.374 ± 0.779 | 6.481 ± 0.617 |
6.744 ± 0.819 | 7.06 ± 0.719 |
1.528 ± 0.266 | 3.635 ± 0.475 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
