
Desulfobacteraceae bacterium SEEP-SAG10
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; unclassified Desulfobacteraceae
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
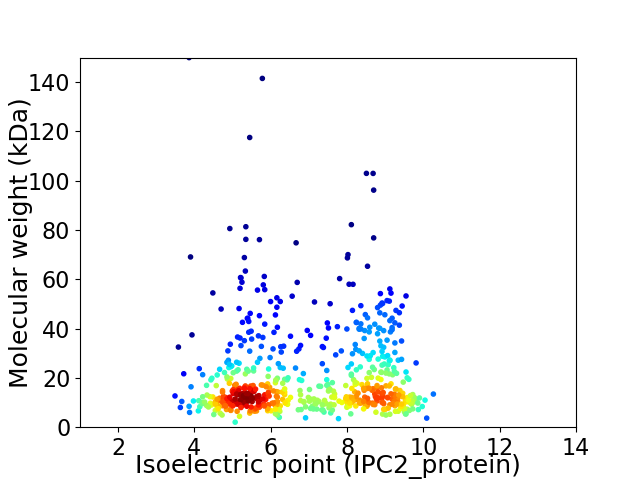
Virtual 2D-PAGE plot for 614 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
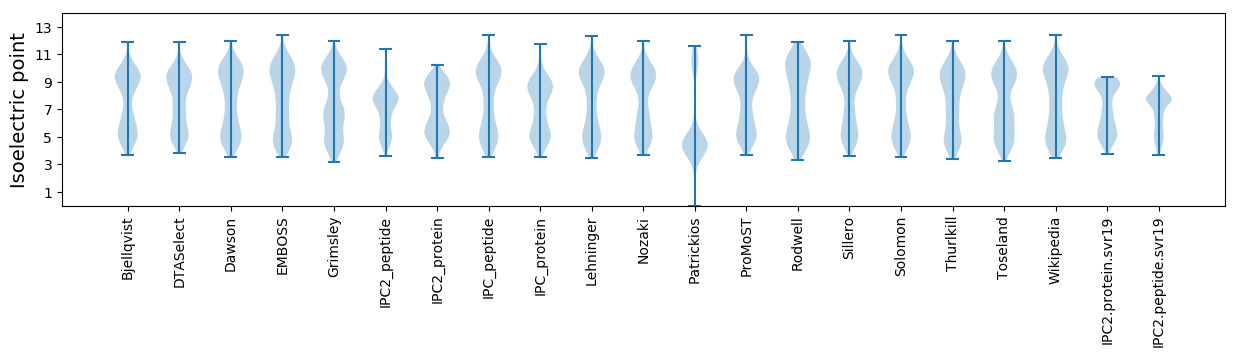
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P6HIT2|A0A2P6HIT2_9DELT Uncharacterized protein OS=Desulfobacteraceae bacterium SEEP-SAG10 OX=2100160 GN=C6A36_00030 PE=4 SV=1
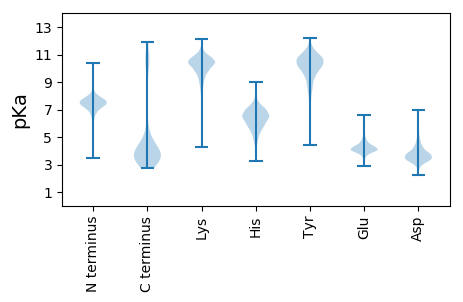
MM1 pKa = 6.82NTSKK5 pKa = 11.09KK6 pKa = 9.52EE7 pKa = 3.94DD8 pKa = 3.09CKK10 pKa = 10.8MFYY13 pKa = 10.56EE14 pKa = 5.02KK15 pKa = 10.96YY16 pKa = 10.28KK17 pKa = 11.12NGDD20 pKa = 3.0NFLINRR26 pKa = 11.84LKK28 pKa = 10.81IMFQFIVIVVLLTSYY43 pKa = 10.98NNSYY47 pKa = 10.35GKK49 pKa = 9.56IWTYY53 pKa = 11.01RR54 pKa = 11.84QITDD58 pKa = 3.54NDD60 pKa = 3.63IKK62 pKa = 11.0DD63 pKa = 3.65SSVSIDD69 pKa = 3.13NGQIAWRR76 pKa = 11.84GYY78 pKa = 9.91GEE80 pKa = 4.05NGKK83 pKa = 9.28LAIFFMDD90 pKa = 3.77LNEE93 pKa = 4.25GVKK96 pKa = 10.65EE97 pKa = 3.9NLTADD102 pKa = 3.26KK103 pKa = 10.9VYY105 pKa = 10.65GYY107 pKa = 10.39FGTPQISNGQVTWRR121 pKa = 11.84RR122 pKa = 11.84YY123 pKa = 10.49DD124 pKa = 3.27GDD126 pKa = 3.87NNKK129 pKa = 10.59DD130 pKa = 4.93SILLWDD136 pKa = 4.52GQDD139 pKa = 2.74IHH141 pKa = 8.18IVSEE145 pKa = 4.27YY146 pKa = 11.0DD147 pKa = 3.62CGDD150 pKa = 3.57FPYY153 pKa = 10.35QGYY156 pKa = 8.04PPSYY160 pKa = 10.65GSLEE164 pKa = 4.1PSLHH168 pKa = 6.23NGQIAYY174 pKa = 9.41AAWDD178 pKa = 3.85GNDD181 pKa = 3.26YY182 pKa = 11.1EE183 pKa = 4.25IFYY186 pKa = 10.72WNGTEE191 pKa = 5.31IIQITDD197 pKa = 3.13NLTNDD202 pKa = 3.85YY203 pKa = 9.4EE204 pKa = 4.26AQLWNGTISYY214 pKa = 8.79TGYY217 pKa = 10.23KK218 pKa = 10.17DD219 pKa = 3.7EE220 pKa = 5.08SAPSDD225 pKa = 3.01IFYY228 pKa = 10.31WDD230 pKa = 3.13GTEE233 pKa = 3.89IQNISDD239 pKa = 3.75KK240 pKa = 11.47AGTCEE245 pKa = 4.15DD246 pKa = 3.18SHH248 pKa = 8.19LRR250 pKa = 11.84EE251 pKa = 4.25SKK253 pKa = 10.06IVHH256 pKa = 5.66SAYY259 pKa = 10.36GLSGEE264 pKa = 4.57YY265 pKa = 9.65GTDD268 pKa = 2.65IYY270 pKa = 10.77IWDD273 pKa = 4.4GSQDD277 pKa = 3.06RR278 pKa = 11.84CLIPVIGYY286 pKa = 9.74DD287 pKa = 3.64YY288 pKa = 10.61EE289 pKa = 4.37PEE291 pKa = 3.91IFEE294 pKa = 6.34SLICWTGWPYY304 pKa = 11.33GKK306 pKa = 9.63PKK308 pKa = 10.04GPCVGWVWDD317 pKa = 4.48GYY319 pKa = 10.72KK320 pKa = 10.19LHH322 pKa = 6.8NVMEE326 pKa = 4.48SPYY329 pKa = 10.4NVSFPRR335 pKa = 11.84ADD337 pKa = 3.13GSMIAFTAHH346 pKa = 7.28DD347 pKa = 4.55GNDD350 pKa = 3.47DD351 pKa = 4.11EE352 pKa = 6.02IFIAEE357 pKa = 4.82YY358 pKa = 8.96ITGTYY363 pKa = 10.23KK364 pKa = 10.66PVTPAGEE371 pKa = 4.17TLNILLEE378 pKa = 4.26NEE380 pKa = 3.98LAYY383 pKa = 10.48VANGEE388 pKa = 4.09GDD390 pKa = 4.07LKK392 pKa = 10.87IVDD395 pKa = 4.26ISDD398 pKa = 3.17EE399 pKa = 4.29GNPVEE404 pKa = 5.27VGGLYY409 pKa = 10.04IPDD412 pKa = 3.79ACSGFEE418 pKa = 3.85VQKK421 pKa = 10.13RR422 pKa = 11.84GNYY425 pKa = 8.61IFLANRR431 pKa = 11.84CGGVKK436 pKa = 9.92IIDD439 pKa = 3.81VTDD442 pKa = 3.52GTNPILVSTVEE453 pKa = 4.21TPDD456 pKa = 3.23MATHH460 pKa = 6.25ITLDD464 pKa = 3.39GNYY467 pKa = 10.25LYY469 pKa = 10.42IGDD472 pKa = 4.18RR473 pKa = 11.84LGGLRR478 pKa = 11.84ILDD481 pKa = 3.47VTIPEE486 pKa = 4.47SVNEE490 pKa = 4.05VGNLDD495 pKa = 4.95LEE497 pKa = 4.51GTTYY501 pKa = 11.17GIAIKK506 pKa = 10.97DD507 pKa = 3.46NFAYY511 pKa = 10.3VANDD515 pKa = 3.17YY516 pKa = 11.51DD517 pKa = 4.51GIQVVDD523 pKa = 3.69ISDD526 pKa = 4.05PEE528 pKa = 4.38SPGMVQRR535 pKa = 11.84YY536 pKa = 8.72DD537 pKa = 3.41LSGITVWDD545 pKa = 3.48IKK547 pKa = 10.9IHH549 pKa = 7.36DD550 pKa = 4.57DD551 pKa = 3.39YY552 pKa = 11.82AYY554 pKa = 10.36LVCPGYY560 pKa = 10.47GIKK563 pKa = 10.53VLDD566 pKa = 3.75ISDD569 pKa = 4.42PYY571 pKa = 11.05DD572 pKa = 3.36LRR574 pKa = 11.84EE575 pKa = 4.09IGEE578 pKa = 4.54CVLPDD583 pKa = 4.03GSRR586 pKa = 11.84PDD588 pKa = 3.84HH589 pKa = 6.5FLPPLDD595 pKa = 3.38ISFVDD600 pKa = 4.17HH601 pKa = 6.23YY602 pKa = 11.81AFVGHH607 pKa = 6.62GNDD610 pKa = 3.61GALVLDD616 pKa = 4.54ISNPYY621 pKa = 8.91YY622 pKa = 9.83PRR624 pKa = 11.84IVEE627 pKa = 4.94RR628 pKa = 11.84IDD630 pKa = 3.28TPGYY634 pKa = 8.7AWGMSIRR641 pKa = 11.84GKK643 pKa = 10.32YY644 pKa = 10.24LYY646 pKa = 10.68VADD649 pKa = 4.61GSEE652 pKa = 4.25GFQIINISDD661 pKa = 3.62YY662 pKa = 10.59MQTLYY667 pKa = 11.11EE668 pKa = 4.33DD669 pKa = 4.79AEE671 pKa = 4.7DD672 pKa = 4.06GRR674 pKa = 11.84ISGWDD679 pKa = 3.16IYY681 pKa = 11.66DD682 pKa = 3.76NSPDD686 pKa = 3.57GAQVTNGFDD695 pKa = 3.47EE696 pKa = 4.91DD697 pKa = 3.68RR698 pKa = 11.84QSRR701 pKa = 11.84VIHH704 pKa = 6.06LNGSGHH710 pKa = 6.51LNGYY714 pKa = 7.64RR715 pKa = 11.84LRR717 pKa = 11.84SDD719 pKa = 5.69DD720 pKa = 3.29GSEE723 pKa = 3.76WHH725 pKa = 6.69NSSQFVIEE733 pKa = 3.87WSMKK737 pKa = 8.06YY738 pKa = 10.32SEE740 pKa = 4.82FFTVYY745 pKa = 10.79VDD747 pKa = 3.55IEE749 pKa = 4.32TTAGQRR755 pKa = 11.84YY756 pKa = 7.02MVYY759 pKa = 10.41KK760 pKa = 10.51PVDD763 pKa = 3.89FDD765 pKa = 4.29DD766 pKa = 5.66LGDD769 pKa = 3.79GGYY772 pKa = 10.23VYY774 pKa = 10.55HH775 pKa = 7.15GLGSDD780 pKa = 3.7TIDD783 pKa = 4.09GQWHH787 pKa = 5.59TFVHH791 pKa = 7.13DD792 pKa = 4.08LQTDD796 pKa = 3.4IEE798 pKa = 4.29EE799 pKa = 4.46AQPGVTILEE808 pKa = 4.23VNGFYY813 pKa = 10.17IRR815 pKa = 11.84GSGSVDD821 pKa = 4.19DD822 pKa = 4.75IGLMDD827 pKa = 4.3TMLINQDD834 pKa = 3.34TDD836 pKa = 3.22SDD838 pKa = 4.81GITDD842 pKa = 4.04FNEE845 pKa = 3.76TNIYY849 pKa = 8.55GTDD852 pKa = 3.62PNKK855 pKa = 10.85ADD857 pKa = 3.74TDD859 pKa = 3.57EE860 pKa = 5.97DD861 pKa = 4.77GIIDD865 pKa = 4.4GDD867 pKa = 3.89EE868 pKa = 3.73LSYY871 pKa = 10.53WGLNWDD877 pKa = 3.88EE878 pKa = 6.03DD879 pKa = 4.49YY880 pKa = 11.69DD881 pKa = 4.42GDD883 pKa = 4.77NLINLLDD890 pKa = 4.64PDD892 pKa = 4.33ADD894 pKa = 3.61NDD896 pKa = 3.88GFLDD900 pKa = 3.97GEE902 pKa = 4.4EE903 pKa = 4.12IAAGSDD909 pKa = 3.4PADD912 pKa = 4.18PEE914 pKa = 4.38SSPLSLVVYY923 pKa = 10.08EE924 pKa = 4.66DD925 pKa = 4.9AEE927 pKa = 4.44DD928 pKa = 3.64SSIIGWDD935 pKa = 3.65VYY937 pKa = 11.73DD938 pKa = 5.6DD939 pKa = 4.93DD940 pKa = 5.49PPGALISNVYY950 pKa = 10.26DD951 pKa = 3.48EE952 pKa = 4.88NRR954 pKa = 11.84QSRR957 pKa = 11.84VIEE960 pKa = 4.11FSGSGVTNGYY970 pKa = 9.16RR971 pKa = 11.84LRR973 pKa = 11.84DD974 pKa = 3.49QNGAPWHH981 pKa = 5.28NTRR984 pKa = 11.84QFVIEE989 pKa = 4.0WSMKK993 pKa = 9.71FDD995 pKa = 3.71QNTNIYY1001 pKa = 10.15VDD1003 pKa = 3.56VEE1005 pKa = 4.5TTSGHH1010 pKa = 6.83RR1011 pKa = 11.84YY1012 pKa = 9.52LLYY1015 pKa = 10.64TPVDD1019 pKa = 3.66YY1020 pKa = 11.24DD1021 pKa = 3.91GLGTGYY1027 pKa = 8.48TVHH1030 pKa = 6.99HH1031 pKa = 6.58GLGVDD1036 pKa = 3.78AKK1038 pKa = 10.3NGLWHH1043 pKa = 6.67TYY1045 pKa = 10.14RR1046 pKa = 11.84RR1047 pKa = 11.84DD1048 pKa = 3.38LQADD1052 pKa = 3.97LEE1054 pKa = 4.39EE1055 pKa = 4.67GQPGVSILEE1064 pKa = 4.11VNGFLIRR1071 pKa = 11.84GSGSVDD1077 pKa = 4.19DD1078 pKa = 4.75IGLMDD1083 pKa = 4.3TMLINQDD1090 pKa = 3.34TDD1092 pKa = 3.22SDD1094 pKa = 4.81GITDD1098 pKa = 4.04FNEE1101 pKa = 3.76TNIYY1105 pKa = 8.55GTDD1108 pKa = 3.62PNKK1111 pKa = 10.85ADD1113 pKa = 3.74TDD1115 pKa = 3.57EE1116 pKa = 5.97DD1117 pKa = 4.77GIIDD1121 pKa = 4.4GDD1123 pKa = 3.89EE1124 pKa = 3.73LSYY1127 pKa = 10.53WGLNWDD1133 pKa = 3.88EE1134 pKa = 6.03DD1135 pKa = 4.49YY1136 pKa = 11.69DD1137 pKa = 4.42GDD1139 pKa = 4.77NLINLLDD1146 pKa = 4.64PDD1148 pKa = 4.33ADD1150 pKa = 3.61NDD1152 pKa = 3.88GFLDD1156 pKa = 3.97GEE1158 pKa = 4.4EE1159 pKa = 4.12IAAGSDD1165 pKa = 3.4PADD1168 pKa = 4.18PEE1170 pKa = 4.38SSPLSLVVYY1179 pKa = 10.08EE1180 pKa = 4.66DD1181 pKa = 4.9AEE1183 pKa = 4.44DD1184 pKa = 3.64SSIIGWDD1191 pKa = 3.65VYY1193 pKa = 11.73DD1194 pKa = 5.6DD1195 pKa = 4.93DD1196 pKa = 5.49PPGALISNVYY1206 pKa = 10.26DD1207 pKa = 3.48EE1208 pKa = 4.88NRR1210 pKa = 11.84QSRR1213 pKa = 11.84VIEE1216 pKa = 4.11FSGSGVTNGYY1226 pKa = 9.16RR1227 pKa = 11.84LRR1229 pKa = 11.84DD1230 pKa = 3.49QNGAPWHH1237 pKa = 5.28NTRR1240 pKa = 11.84QFVIEE1245 pKa = 4.0WSMKK1249 pKa = 9.71FDD1251 pKa = 3.71QNTNIYY1257 pKa = 10.15VDD1259 pKa = 3.56VEE1261 pKa = 4.5TTSGHH1266 pKa = 6.83RR1267 pKa = 11.84YY1268 pKa = 9.52LLYY1271 pKa = 10.64TPVDD1275 pKa = 3.66YY1276 pKa = 11.24DD1277 pKa = 3.91GLGTGYY1283 pKa = 8.48TVHH1286 pKa = 6.99HH1287 pKa = 6.58GLGVDD1292 pKa = 3.78AKK1294 pKa = 10.3NGLWHH1299 pKa = 6.67TYY1301 pKa = 10.14RR1302 pKa = 11.84RR1303 pKa = 11.84DD1304 pKa = 3.38LQADD1308 pKa = 3.97LEE1310 pKa = 4.39EE1311 pKa = 4.67GQPGVSILEE1320 pKa = 4.11VNGFLIRR1327 pKa = 11.84GSGSLDD1333 pKa = 3.92DD1334 pKa = 5.71IILKK1338 pKa = 9.14EE1339 pKa = 4.09QYY1341 pKa = 10.63
MM1 pKa = 6.82NTSKK5 pKa = 11.09KK6 pKa = 9.52EE7 pKa = 3.94DD8 pKa = 3.09CKK10 pKa = 10.8MFYY13 pKa = 10.56EE14 pKa = 5.02KK15 pKa = 10.96YY16 pKa = 10.28KK17 pKa = 11.12NGDD20 pKa = 3.0NFLINRR26 pKa = 11.84LKK28 pKa = 10.81IMFQFIVIVVLLTSYY43 pKa = 10.98NNSYY47 pKa = 10.35GKK49 pKa = 9.56IWTYY53 pKa = 11.01RR54 pKa = 11.84QITDD58 pKa = 3.54NDD60 pKa = 3.63IKK62 pKa = 11.0DD63 pKa = 3.65SSVSIDD69 pKa = 3.13NGQIAWRR76 pKa = 11.84GYY78 pKa = 9.91GEE80 pKa = 4.05NGKK83 pKa = 9.28LAIFFMDD90 pKa = 3.77LNEE93 pKa = 4.25GVKK96 pKa = 10.65EE97 pKa = 3.9NLTADD102 pKa = 3.26KK103 pKa = 10.9VYY105 pKa = 10.65GYY107 pKa = 10.39FGTPQISNGQVTWRR121 pKa = 11.84RR122 pKa = 11.84YY123 pKa = 10.49DD124 pKa = 3.27GDD126 pKa = 3.87NNKK129 pKa = 10.59DD130 pKa = 4.93SILLWDD136 pKa = 4.52GQDD139 pKa = 2.74IHH141 pKa = 8.18IVSEE145 pKa = 4.27YY146 pKa = 11.0DD147 pKa = 3.62CGDD150 pKa = 3.57FPYY153 pKa = 10.35QGYY156 pKa = 8.04PPSYY160 pKa = 10.65GSLEE164 pKa = 4.1PSLHH168 pKa = 6.23NGQIAYY174 pKa = 9.41AAWDD178 pKa = 3.85GNDD181 pKa = 3.26YY182 pKa = 11.1EE183 pKa = 4.25IFYY186 pKa = 10.72WNGTEE191 pKa = 5.31IIQITDD197 pKa = 3.13NLTNDD202 pKa = 3.85YY203 pKa = 9.4EE204 pKa = 4.26AQLWNGTISYY214 pKa = 8.79TGYY217 pKa = 10.23KK218 pKa = 10.17DD219 pKa = 3.7EE220 pKa = 5.08SAPSDD225 pKa = 3.01IFYY228 pKa = 10.31WDD230 pKa = 3.13GTEE233 pKa = 3.89IQNISDD239 pKa = 3.75KK240 pKa = 11.47AGTCEE245 pKa = 4.15DD246 pKa = 3.18SHH248 pKa = 8.19LRR250 pKa = 11.84EE251 pKa = 4.25SKK253 pKa = 10.06IVHH256 pKa = 5.66SAYY259 pKa = 10.36GLSGEE264 pKa = 4.57YY265 pKa = 9.65GTDD268 pKa = 2.65IYY270 pKa = 10.77IWDD273 pKa = 4.4GSQDD277 pKa = 3.06RR278 pKa = 11.84CLIPVIGYY286 pKa = 9.74DD287 pKa = 3.64YY288 pKa = 10.61EE289 pKa = 4.37PEE291 pKa = 3.91IFEE294 pKa = 6.34SLICWTGWPYY304 pKa = 11.33GKK306 pKa = 9.63PKK308 pKa = 10.04GPCVGWVWDD317 pKa = 4.48GYY319 pKa = 10.72KK320 pKa = 10.19LHH322 pKa = 6.8NVMEE326 pKa = 4.48SPYY329 pKa = 10.4NVSFPRR335 pKa = 11.84ADD337 pKa = 3.13GSMIAFTAHH346 pKa = 7.28DD347 pKa = 4.55GNDD350 pKa = 3.47DD351 pKa = 4.11EE352 pKa = 6.02IFIAEE357 pKa = 4.82YY358 pKa = 8.96ITGTYY363 pKa = 10.23KK364 pKa = 10.66PVTPAGEE371 pKa = 4.17TLNILLEE378 pKa = 4.26NEE380 pKa = 3.98LAYY383 pKa = 10.48VANGEE388 pKa = 4.09GDD390 pKa = 4.07LKK392 pKa = 10.87IVDD395 pKa = 4.26ISDD398 pKa = 3.17EE399 pKa = 4.29GNPVEE404 pKa = 5.27VGGLYY409 pKa = 10.04IPDD412 pKa = 3.79ACSGFEE418 pKa = 3.85VQKK421 pKa = 10.13RR422 pKa = 11.84GNYY425 pKa = 8.61IFLANRR431 pKa = 11.84CGGVKK436 pKa = 9.92IIDD439 pKa = 3.81VTDD442 pKa = 3.52GTNPILVSTVEE453 pKa = 4.21TPDD456 pKa = 3.23MATHH460 pKa = 6.25ITLDD464 pKa = 3.39GNYY467 pKa = 10.25LYY469 pKa = 10.42IGDD472 pKa = 4.18RR473 pKa = 11.84LGGLRR478 pKa = 11.84ILDD481 pKa = 3.47VTIPEE486 pKa = 4.47SVNEE490 pKa = 4.05VGNLDD495 pKa = 4.95LEE497 pKa = 4.51GTTYY501 pKa = 11.17GIAIKK506 pKa = 10.97DD507 pKa = 3.46NFAYY511 pKa = 10.3VANDD515 pKa = 3.17YY516 pKa = 11.51DD517 pKa = 4.51GIQVVDD523 pKa = 3.69ISDD526 pKa = 4.05PEE528 pKa = 4.38SPGMVQRR535 pKa = 11.84YY536 pKa = 8.72DD537 pKa = 3.41LSGITVWDD545 pKa = 3.48IKK547 pKa = 10.9IHH549 pKa = 7.36DD550 pKa = 4.57DD551 pKa = 3.39YY552 pKa = 11.82AYY554 pKa = 10.36LVCPGYY560 pKa = 10.47GIKK563 pKa = 10.53VLDD566 pKa = 3.75ISDD569 pKa = 4.42PYY571 pKa = 11.05DD572 pKa = 3.36LRR574 pKa = 11.84EE575 pKa = 4.09IGEE578 pKa = 4.54CVLPDD583 pKa = 4.03GSRR586 pKa = 11.84PDD588 pKa = 3.84HH589 pKa = 6.5FLPPLDD595 pKa = 3.38ISFVDD600 pKa = 4.17HH601 pKa = 6.23YY602 pKa = 11.81AFVGHH607 pKa = 6.62GNDD610 pKa = 3.61GALVLDD616 pKa = 4.54ISNPYY621 pKa = 8.91YY622 pKa = 9.83PRR624 pKa = 11.84IVEE627 pKa = 4.94RR628 pKa = 11.84IDD630 pKa = 3.28TPGYY634 pKa = 8.7AWGMSIRR641 pKa = 11.84GKK643 pKa = 10.32YY644 pKa = 10.24LYY646 pKa = 10.68VADD649 pKa = 4.61GSEE652 pKa = 4.25GFQIINISDD661 pKa = 3.62YY662 pKa = 10.59MQTLYY667 pKa = 11.11EE668 pKa = 4.33DD669 pKa = 4.79AEE671 pKa = 4.7DD672 pKa = 4.06GRR674 pKa = 11.84ISGWDD679 pKa = 3.16IYY681 pKa = 11.66DD682 pKa = 3.76NSPDD686 pKa = 3.57GAQVTNGFDD695 pKa = 3.47EE696 pKa = 4.91DD697 pKa = 3.68RR698 pKa = 11.84QSRR701 pKa = 11.84VIHH704 pKa = 6.06LNGSGHH710 pKa = 6.51LNGYY714 pKa = 7.64RR715 pKa = 11.84LRR717 pKa = 11.84SDD719 pKa = 5.69DD720 pKa = 3.29GSEE723 pKa = 3.76WHH725 pKa = 6.69NSSQFVIEE733 pKa = 3.87WSMKK737 pKa = 8.06YY738 pKa = 10.32SEE740 pKa = 4.82FFTVYY745 pKa = 10.79VDD747 pKa = 3.55IEE749 pKa = 4.32TTAGQRR755 pKa = 11.84YY756 pKa = 7.02MVYY759 pKa = 10.41KK760 pKa = 10.51PVDD763 pKa = 3.89FDD765 pKa = 4.29DD766 pKa = 5.66LGDD769 pKa = 3.79GGYY772 pKa = 10.23VYY774 pKa = 10.55HH775 pKa = 7.15GLGSDD780 pKa = 3.7TIDD783 pKa = 4.09GQWHH787 pKa = 5.59TFVHH791 pKa = 7.13DD792 pKa = 4.08LQTDD796 pKa = 3.4IEE798 pKa = 4.29EE799 pKa = 4.46AQPGVTILEE808 pKa = 4.23VNGFYY813 pKa = 10.17IRR815 pKa = 11.84GSGSVDD821 pKa = 4.19DD822 pKa = 4.75IGLMDD827 pKa = 4.3TMLINQDD834 pKa = 3.34TDD836 pKa = 3.22SDD838 pKa = 4.81GITDD842 pKa = 4.04FNEE845 pKa = 3.76TNIYY849 pKa = 8.55GTDD852 pKa = 3.62PNKK855 pKa = 10.85ADD857 pKa = 3.74TDD859 pKa = 3.57EE860 pKa = 5.97DD861 pKa = 4.77GIIDD865 pKa = 4.4GDD867 pKa = 3.89EE868 pKa = 3.73LSYY871 pKa = 10.53WGLNWDD877 pKa = 3.88EE878 pKa = 6.03DD879 pKa = 4.49YY880 pKa = 11.69DD881 pKa = 4.42GDD883 pKa = 4.77NLINLLDD890 pKa = 4.64PDD892 pKa = 4.33ADD894 pKa = 3.61NDD896 pKa = 3.88GFLDD900 pKa = 3.97GEE902 pKa = 4.4EE903 pKa = 4.12IAAGSDD909 pKa = 3.4PADD912 pKa = 4.18PEE914 pKa = 4.38SSPLSLVVYY923 pKa = 10.08EE924 pKa = 4.66DD925 pKa = 4.9AEE927 pKa = 4.44DD928 pKa = 3.64SSIIGWDD935 pKa = 3.65VYY937 pKa = 11.73DD938 pKa = 5.6DD939 pKa = 4.93DD940 pKa = 5.49PPGALISNVYY950 pKa = 10.26DD951 pKa = 3.48EE952 pKa = 4.88NRR954 pKa = 11.84QSRR957 pKa = 11.84VIEE960 pKa = 4.11FSGSGVTNGYY970 pKa = 9.16RR971 pKa = 11.84LRR973 pKa = 11.84DD974 pKa = 3.49QNGAPWHH981 pKa = 5.28NTRR984 pKa = 11.84QFVIEE989 pKa = 4.0WSMKK993 pKa = 9.71FDD995 pKa = 3.71QNTNIYY1001 pKa = 10.15VDD1003 pKa = 3.56VEE1005 pKa = 4.5TTSGHH1010 pKa = 6.83RR1011 pKa = 11.84YY1012 pKa = 9.52LLYY1015 pKa = 10.64TPVDD1019 pKa = 3.66YY1020 pKa = 11.24DD1021 pKa = 3.91GLGTGYY1027 pKa = 8.48TVHH1030 pKa = 6.99HH1031 pKa = 6.58GLGVDD1036 pKa = 3.78AKK1038 pKa = 10.3NGLWHH1043 pKa = 6.67TYY1045 pKa = 10.14RR1046 pKa = 11.84RR1047 pKa = 11.84DD1048 pKa = 3.38LQADD1052 pKa = 3.97LEE1054 pKa = 4.39EE1055 pKa = 4.67GQPGVSILEE1064 pKa = 4.11VNGFLIRR1071 pKa = 11.84GSGSVDD1077 pKa = 4.19DD1078 pKa = 4.75IGLMDD1083 pKa = 4.3TMLINQDD1090 pKa = 3.34TDD1092 pKa = 3.22SDD1094 pKa = 4.81GITDD1098 pKa = 4.04FNEE1101 pKa = 3.76TNIYY1105 pKa = 8.55GTDD1108 pKa = 3.62PNKK1111 pKa = 10.85ADD1113 pKa = 3.74TDD1115 pKa = 3.57EE1116 pKa = 5.97DD1117 pKa = 4.77GIIDD1121 pKa = 4.4GDD1123 pKa = 3.89EE1124 pKa = 3.73LSYY1127 pKa = 10.53WGLNWDD1133 pKa = 3.88EE1134 pKa = 6.03DD1135 pKa = 4.49YY1136 pKa = 11.69DD1137 pKa = 4.42GDD1139 pKa = 4.77NLINLLDD1146 pKa = 4.64PDD1148 pKa = 4.33ADD1150 pKa = 3.61NDD1152 pKa = 3.88GFLDD1156 pKa = 3.97GEE1158 pKa = 4.4EE1159 pKa = 4.12IAAGSDD1165 pKa = 3.4PADD1168 pKa = 4.18PEE1170 pKa = 4.38SSPLSLVVYY1179 pKa = 10.08EE1180 pKa = 4.66DD1181 pKa = 4.9AEE1183 pKa = 4.44DD1184 pKa = 3.64SSIIGWDD1191 pKa = 3.65VYY1193 pKa = 11.73DD1194 pKa = 5.6DD1195 pKa = 4.93DD1196 pKa = 5.49PPGALISNVYY1206 pKa = 10.26DD1207 pKa = 3.48EE1208 pKa = 4.88NRR1210 pKa = 11.84QSRR1213 pKa = 11.84VIEE1216 pKa = 4.11FSGSGVTNGYY1226 pKa = 9.16RR1227 pKa = 11.84LRR1229 pKa = 11.84DD1230 pKa = 3.49QNGAPWHH1237 pKa = 5.28NTRR1240 pKa = 11.84QFVIEE1245 pKa = 4.0WSMKK1249 pKa = 9.71FDD1251 pKa = 3.71QNTNIYY1257 pKa = 10.15VDD1259 pKa = 3.56VEE1261 pKa = 4.5TTSGHH1266 pKa = 6.83RR1267 pKa = 11.84YY1268 pKa = 9.52LLYY1271 pKa = 10.64TPVDD1275 pKa = 3.66YY1276 pKa = 11.24DD1277 pKa = 3.91GLGTGYY1283 pKa = 8.48TVHH1286 pKa = 6.99HH1287 pKa = 6.58GLGVDD1292 pKa = 3.78AKK1294 pKa = 10.3NGLWHH1299 pKa = 6.67TYY1301 pKa = 10.14RR1302 pKa = 11.84RR1303 pKa = 11.84DD1304 pKa = 3.38LQADD1308 pKa = 3.97LEE1310 pKa = 4.39EE1311 pKa = 4.67GQPGVSILEE1320 pKa = 4.11VNGFLIRR1327 pKa = 11.84GSGSLDD1333 pKa = 3.92DD1334 pKa = 5.71IILKK1338 pKa = 9.14EE1339 pKa = 4.09QYY1341 pKa = 10.63
Molecular weight: 149.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P6HHA8|A0A2P6HHA8_9DELT Arginine--tRNA ligase (Fragment) OS=Desulfobacteraceae bacterium SEEP-SAG10 OX=2100160 GN=argS PE=3 SV=1
LL1 pKa = 7.12LRR3 pKa = 11.84YY4 pKa = 9.76GDD6 pKa = 4.1RR7 pKa = 11.84VNMAFGVEE15 pKa = 4.11NRR17 pKa = 11.84CPFLDD22 pKa = 3.36HH23 pKa = 7.29RR24 pKa = 11.84LIEE27 pKa = 4.31YY28 pKa = 10.4VSALPSHH35 pKa = 5.97MKK37 pKa = 9.95IRR39 pKa = 11.84GGTTKK44 pKa = 10.08WIFRR48 pKa = 11.84EE49 pKa = 4.18VADD52 pKa = 4.06GRR54 pKa = 11.84IPKK57 pKa = 10.2AILKK61 pKa = 10.02RR62 pKa = 11.84RR63 pKa = 11.84LKK65 pKa = 10.32MGFPTPLGVWLRR77 pKa = 11.84KK78 pKa = 9.02EE79 pKa = 4.03LPEE82 pKa = 5.38VARR85 pKa = 11.84QWLGNYY91 pKa = 9.26RR92 pKa = 11.84FLPFFDD98 pKa = 3.31RR99 pKa = 11.84WII101 pKa = 4.14
LL1 pKa = 7.12LRR3 pKa = 11.84YY4 pKa = 9.76GDD6 pKa = 4.1RR7 pKa = 11.84VNMAFGVEE15 pKa = 4.11NRR17 pKa = 11.84CPFLDD22 pKa = 3.36HH23 pKa = 7.29RR24 pKa = 11.84LIEE27 pKa = 4.31YY28 pKa = 10.4VSALPSHH35 pKa = 5.97MKK37 pKa = 9.95IRR39 pKa = 11.84GGTTKK44 pKa = 10.08WIFRR48 pKa = 11.84EE49 pKa = 4.18VADD52 pKa = 4.06GRR54 pKa = 11.84IPKK57 pKa = 10.2AILKK61 pKa = 10.02RR62 pKa = 11.84RR63 pKa = 11.84LKK65 pKa = 10.32MGFPTPLGVWLRR77 pKa = 11.84KK78 pKa = 9.02EE79 pKa = 4.03LPEE82 pKa = 5.38VARR85 pKa = 11.84QWLGNYY91 pKa = 9.26RR92 pKa = 11.84FLPFFDD98 pKa = 3.31RR99 pKa = 11.84WII101 pKa = 4.14
Molecular weight: 12.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
116105 |
20 |
1341 |
189.1 |
21.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.695 ± 0.117 | 1.273 ± 0.046 |
5.704 ± 0.114 | 6.492 ± 0.1 |
4.621 ± 0.092 | 6.739 ± 0.13 |
2.117 ± 0.051 | 7.935 ± 0.116 |
7.502 ± 0.144 | 9.465 ± 0.123 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.476 ± 0.051 | 4.39 ± 0.079 |
4.043 ± 0.061 | 3.168 ± 0.069 |
5.296 ± 0.092 | 6.15 ± 0.083 |
5.038 ± 0.075 | 6.324 ± 0.093 |
1.111 ± 0.051 | 3.462 ± 0.073 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
