
Mesonia algae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes;
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
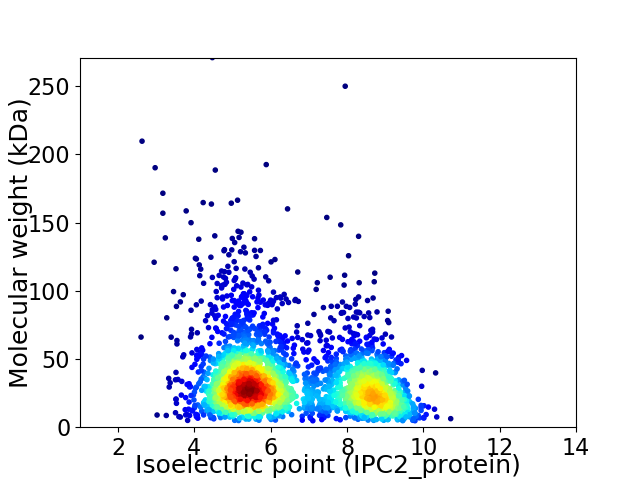
Virtual 2D-PAGE plot for 2856 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2W7HW89|A0A2W7HW89_9FLAO DNA helicase OS=Mesonia algae OX=213248 GN=LX95_02870 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 9.28KK3 pKa = 8.54TYY5 pKa = 10.72LSILIALISITSFAQEE21 pKa = 3.96ALITGYY27 pKa = 10.66VDD29 pKa = 4.1SPCSGANGRR38 pKa = 11.84VLEE41 pKa = 4.38IYY43 pKa = 11.17VNGTIDD49 pKa = 3.52FTGWNIQRR57 pKa = 11.84QSNGGGYY64 pKa = 7.78STNIDD69 pKa = 3.49LSSFATISDD78 pKa = 3.52DD79 pKa = 5.77FIYY82 pKa = 9.52LTNDD86 pKa = 3.11QAILNAEE93 pKa = 4.19FGITANVIEE102 pKa = 4.62SSVISSNGNDD112 pKa = 3.16AFQIIDD118 pKa = 3.74NNSVVIDD125 pKa = 3.76RR126 pKa = 11.84FGEE129 pKa = 4.1EE130 pKa = 4.37NIDD133 pKa = 3.71GAGTAWEE140 pKa = 4.68HH141 pKa = 6.37LDD143 pKa = 4.52SYY145 pKa = 9.99YY146 pKa = 11.02LRR148 pKa = 11.84NNSSIANAGSFDD160 pKa = 3.79VNNWTFGALNSLDD173 pKa = 3.71NQGLCNSSSALSTMVNLGSFTPVVTTVAEE202 pKa = 4.14IEE204 pKa = 3.94FDD206 pKa = 3.26EE207 pKa = 5.34AYY209 pKa = 9.78ISAVEE214 pKa = 4.22GSGTATLSVTISEE227 pKa = 5.02LPNSDD232 pKa = 3.88ANIDD236 pKa = 3.58VAILVNEE243 pKa = 4.08STAIDD248 pKa = 3.9NQHH251 pKa = 4.75YY252 pKa = 8.7TYY254 pKa = 10.9TNEE257 pKa = 4.0TLTFTTTGSLSQSITITIPDD277 pKa = 3.93NFDD280 pKa = 3.58AEE282 pKa = 4.61ADD284 pKa = 3.68TFLALEE290 pKa = 4.23LTNPVNANLGEE301 pKa = 4.79DD302 pKa = 3.73NLSVVYY308 pKa = 10.56ILDD311 pKa = 4.01NEE313 pKa = 4.26AHH315 pKa = 6.44SPIATQNLDD324 pKa = 3.21INFATSYY331 pKa = 10.88AITGPNPGSEE341 pKa = 3.89IVAHH345 pKa = 6.86DD346 pKa = 3.74ATTEE350 pKa = 3.89RR351 pKa = 11.84LFVMNSGNASVEE363 pKa = 3.91ILDD366 pKa = 4.23FSSPLNITSISTIDD380 pKa = 3.45LSSYY384 pKa = 10.61GDD386 pKa = 3.33SSTSVAVHH394 pKa = 6.38NGLVAATAVPNDD406 pKa = 3.34KK407 pKa = 10.66NLNGKK412 pKa = 7.96VVFMDD417 pKa = 3.52TNGVIITSLEE427 pKa = 4.21VGSLPDD433 pKa = 4.48MITFSPDD440 pKa = 2.53GNMLLVANEE449 pKa = 4.39GEE451 pKa = 4.3PNSDD455 pKa = 3.33YY456 pKa = 11.21TIDD459 pKa = 3.77PEE461 pKa = 4.49GTISIIDD468 pKa = 3.78LSIATANLTQANVTTLNFNAFDD490 pKa = 3.83TQEE493 pKa = 4.21TQLKK497 pKa = 9.62ADD499 pKa = 3.94GVRR502 pKa = 11.84IFGPNATVSQDD513 pKa = 3.74LEE515 pKa = 4.14PEE517 pKa = 4.42YY518 pKa = 10.37ITVSQDD524 pKa = 2.49NTTAWVSLQEE534 pKa = 4.06NNAIAVVDD542 pKa = 4.43LTTMQITNILPLGLKK557 pKa = 10.03DD558 pKa = 4.43HH559 pKa = 6.77SLPNNALDD567 pKa = 3.84TSNEE571 pKa = 3.49QDD573 pKa = 4.31FIFMANWPIKK583 pKa = 9.56GMYY586 pKa = 8.77MPDD589 pKa = 4.13AIASYY594 pKa = 10.4SVNGNTYY601 pKa = 9.88IVTANEE607 pKa = 3.7GDD609 pKa = 3.3ARR611 pKa = 11.84EE612 pKa = 3.91YY613 pKa = 10.67DD614 pKa = 3.6TFEE617 pKa = 4.23EE618 pKa = 4.52EE619 pKa = 4.79VNIEE623 pKa = 4.74DD624 pKa = 3.55IVLDD628 pKa = 3.68ASVFPNQDD636 pKa = 3.06FLEE639 pKa = 4.97LDD641 pKa = 3.7EE642 pKa = 4.99NLGKK646 pKa = 10.31IKK648 pKa = 9.35ITSEE652 pKa = 4.02FGDD655 pKa = 3.45IDD657 pKa = 3.88NDD659 pKa = 4.22GEE661 pKa = 4.13FEE663 pKa = 4.31EE664 pKa = 4.39IHH666 pKa = 6.1VFGGRR671 pKa = 11.84SFSIYY676 pKa = 10.51DD677 pKa = 4.77AITGAQVYY685 pKa = 10.54DD686 pKa = 3.59SGSDD690 pKa = 3.42FEE692 pKa = 5.74RR693 pKa = 11.84IIAEE697 pKa = 4.06DD698 pKa = 3.55PVYY701 pKa = 10.99NAIFNATDD709 pKa = 4.27DD710 pKa = 3.99EE711 pKa = 4.8NEE713 pKa = 4.08LKK715 pKa = 10.75NRR717 pKa = 11.84SDD719 pKa = 3.58NKK721 pKa = 10.23GPEE724 pKa = 3.98PEE726 pKa = 3.86AVIVQEE732 pKa = 4.3IDD734 pKa = 3.06GTFYY738 pKa = 11.49AFIALEE744 pKa = 4.06RR745 pKa = 11.84VGGFMVYY752 pKa = 10.47DD753 pKa = 3.68ITNPNTPVFEE763 pKa = 5.01GYY765 pKa = 10.42FNNRR769 pKa = 11.84SVTPGEE775 pKa = 3.98GDD777 pKa = 3.51LANLGDD783 pKa = 4.7LAPEE787 pKa = 4.38SIVYY791 pKa = 9.66IAPEE795 pKa = 3.79NNAEE799 pKa = 4.1NKK801 pKa = 10.15GLIVIANEE809 pKa = 3.7VSATISVYY817 pKa = 8.27TLEE820 pKa = 4.51NNVLTTEE827 pKa = 4.04SFEE830 pKa = 4.42VNNSFVVYY838 pKa = 8.74PNPATSSRR846 pKa = 11.84VFFAQPTTYY855 pKa = 10.61EE856 pKa = 4.23LYY858 pKa = 10.44DD859 pKa = 3.27IQGRR863 pKa = 11.84KK864 pKa = 7.94VKK866 pKa = 10.17AAQNSTFINVDD877 pKa = 3.19GLNAGTYY884 pKa = 8.76IVKK887 pKa = 10.35NDD889 pKa = 3.33GGQTQKK895 pKa = 11.4LIIKK899 pKa = 9.66
MM1 pKa = 7.61KK2 pKa = 9.28KK3 pKa = 8.54TYY5 pKa = 10.72LSILIALISITSFAQEE21 pKa = 3.96ALITGYY27 pKa = 10.66VDD29 pKa = 4.1SPCSGANGRR38 pKa = 11.84VLEE41 pKa = 4.38IYY43 pKa = 11.17VNGTIDD49 pKa = 3.52FTGWNIQRR57 pKa = 11.84QSNGGGYY64 pKa = 7.78STNIDD69 pKa = 3.49LSSFATISDD78 pKa = 3.52DD79 pKa = 5.77FIYY82 pKa = 9.52LTNDD86 pKa = 3.11QAILNAEE93 pKa = 4.19FGITANVIEE102 pKa = 4.62SSVISSNGNDD112 pKa = 3.16AFQIIDD118 pKa = 3.74NNSVVIDD125 pKa = 3.76RR126 pKa = 11.84FGEE129 pKa = 4.1EE130 pKa = 4.37NIDD133 pKa = 3.71GAGTAWEE140 pKa = 4.68HH141 pKa = 6.37LDD143 pKa = 4.52SYY145 pKa = 9.99YY146 pKa = 11.02LRR148 pKa = 11.84NNSSIANAGSFDD160 pKa = 3.79VNNWTFGALNSLDD173 pKa = 3.71NQGLCNSSSALSTMVNLGSFTPVVTTVAEE202 pKa = 4.14IEE204 pKa = 3.94FDD206 pKa = 3.26EE207 pKa = 5.34AYY209 pKa = 9.78ISAVEE214 pKa = 4.22GSGTATLSVTISEE227 pKa = 5.02LPNSDD232 pKa = 3.88ANIDD236 pKa = 3.58VAILVNEE243 pKa = 4.08STAIDD248 pKa = 3.9NQHH251 pKa = 4.75YY252 pKa = 8.7TYY254 pKa = 10.9TNEE257 pKa = 4.0TLTFTTTGSLSQSITITIPDD277 pKa = 3.93NFDD280 pKa = 3.58AEE282 pKa = 4.61ADD284 pKa = 3.68TFLALEE290 pKa = 4.23LTNPVNANLGEE301 pKa = 4.79DD302 pKa = 3.73NLSVVYY308 pKa = 10.56ILDD311 pKa = 4.01NEE313 pKa = 4.26AHH315 pKa = 6.44SPIATQNLDD324 pKa = 3.21INFATSYY331 pKa = 10.88AITGPNPGSEE341 pKa = 3.89IVAHH345 pKa = 6.86DD346 pKa = 3.74ATTEE350 pKa = 3.89RR351 pKa = 11.84LFVMNSGNASVEE363 pKa = 3.91ILDD366 pKa = 4.23FSSPLNITSISTIDD380 pKa = 3.45LSSYY384 pKa = 10.61GDD386 pKa = 3.33SSTSVAVHH394 pKa = 6.38NGLVAATAVPNDD406 pKa = 3.34KK407 pKa = 10.66NLNGKK412 pKa = 7.96VVFMDD417 pKa = 3.52TNGVIITSLEE427 pKa = 4.21VGSLPDD433 pKa = 4.48MITFSPDD440 pKa = 2.53GNMLLVANEE449 pKa = 4.39GEE451 pKa = 4.3PNSDD455 pKa = 3.33YY456 pKa = 11.21TIDD459 pKa = 3.77PEE461 pKa = 4.49GTISIIDD468 pKa = 3.78LSIATANLTQANVTTLNFNAFDD490 pKa = 3.83TQEE493 pKa = 4.21TQLKK497 pKa = 9.62ADD499 pKa = 3.94GVRR502 pKa = 11.84IFGPNATVSQDD513 pKa = 3.74LEE515 pKa = 4.14PEE517 pKa = 4.42YY518 pKa = 10.37ITVSQDD524 pKa = 2.49NTTAWVSLQEE534 pKa = 4.06NNAIAVVDD542 pKa = 4.43LTTMQITNILPLGLKK557 pKa = 10.03DD558 pKa = 4.43HH559 pKa = 6.77SLPNNALDD567 pKa = 3.84TSNEE571 pKa = 3.49QDD573 pKa = 4.31FIFMANWPIKK583 pKa = 9.56GMYY586 pKa = 8.77MPDD589 pKa = 4.13AIASYY594 pKa = 10.4SVNGNTYY601 pKa = 9.88IVTANEE607 pKa = 3.7GDD609 pKa = 3.3ARR611 pKa = 11.84EE612 pKa = 3.91YY613 pKa = 10.67DD614 pKa = 3.6TFEE617 pKa = 4.23EE618 pKa = 4.52EE619 pKa = 4.79VNIEE623 pKa = 4.74DD624 pKa = 3.55IVLDD628 pKa = 3.68ASVFPNQDD636 pKa = 3.06FLEE639 pKa = 4.97LDD641 pKa = 3.7EE642 pKa = 4.99NLGKK646 pKa = 10.31IKK648 pKa = 9.35ITSEE652 pKa = 4.02FGDD655 pKa = 3.45IDD657 pKa = 3.88NDD659 pKa = 4.22GEE661 pKa = 4.13FEE663 pKa = 4.31EE664 pKa = 4.39IHH666 pKa = 6.1VFGGRR671 pKa = 11.84SFSIYY676 pKa = 10.51DD677 pKa = 4.77AITGAQVYY685 pKa = 10.54DD686 pKa = 3.59SGSDD690 pKa = 3.42FEE692 pKa = 5.74RR693 pKa = 11.84IIAEE697 pKa = 4.06DD698 pKa = 3.55PVYY701 pKa = 10.99NAIFNATDD709 pKa = 4.27DD710 pKa = 3.99EE711 pKa = 4.8NEE713 pKa = 4.08LKK715 pKa = 10.75NRR717 pKa = 11.84SDD719 pKa = 3.58NKK721 pKa = 10.23GPEE724 pKa = 3.98PEE726 pKa = 3.86AVIVQEE732 pKa = 4.3IDD734 pKa = 3.06GTFYY738 pKa = 11.49AFIALEE744 pKa = 4.06RR745 pKa = 11.84VGGFMVYY752 pKa = 10.47DD753 pKa = 3.68ITNPNTPVFEE763 pKa = 5.01GYY765 pKa = 10.42FNNRR769 pKa = 11.84SVTPGEE775 pKa = 3.98GDD777 pKa = 3.51LANLGDD783 pKa = 4.7LAPEE787 pKa = 4.38SIVYY791 pKa = 9.66IAPEE795 pKa = 3.79NNAEE799 pKa = 4.1NKK801 pKa = 10.15GLIVIANEE809 pKa = 3.7VSATISVYY817 pKa = 8.27TLEE820 pKa = 4.51NNVLTTEE827 pKa = 4.04SFEE830 pKa = 4.42VNNSFVVYY838 pKa = 8.74PNPATSSRR846 pKa = 11.84VFFAQPTTYY855 pKa = 10.61EE856 pKa = 4.23LYY858 pKa = 10.44DD859 pKa = 3.27IQGRR863 pKa = 11.84KK864 pKa = 7.94VKK866 pKa = 10.17AAQNSTFINVDD877 pKa = 3.19GLNAGTYY884 pKa = 8.76IVKK887 pKa = 10.35NDD889 pKa = 3.33GGQTQKK895 pKa = 11.4LIIKK899 pKa = 9.66
Molecular weight: 97.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2W7JUA2|A0A2W7JUA2_9FLAO Uncharacterized protein OS=Mesonia algae OX=213248 GN=LX95_02170 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
933224 |
40 |
2403 |
326.8 |
37.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.09 ± 0.041 | 0.71 ± 0.016 |
5.351 ± 0.041 | 7.24 ± 0.046 |
5.262 ± 0.035 | 6.062 ± 0.045 |
1.798 ± 0.024 | 8.009 ± 0.041 |
8.133 ± 0.074 | 9.504 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.103 ± 0.025 | 6.241 ± 0.055 |
3.268 ± 0.022 | 3.763 ± 0.032 |
3.244 ± 0.033 | 6.559 ± 0.042 |
5.569 ± 0.042 | 6.029 ± 0.034 |
0.976 ± 0.018 | 4.09 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
