
Beihai sobemo-like virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
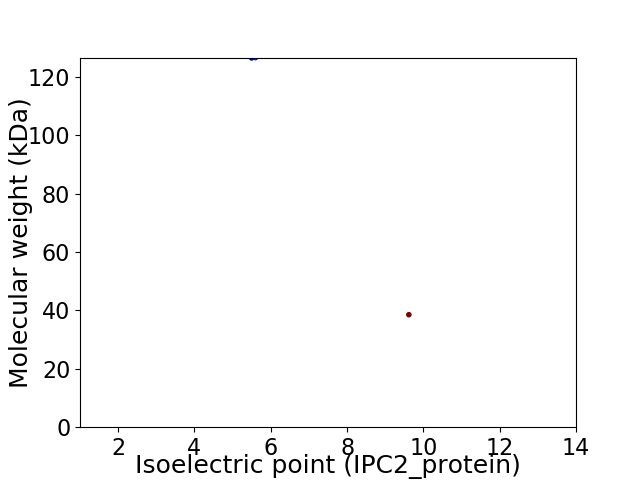
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
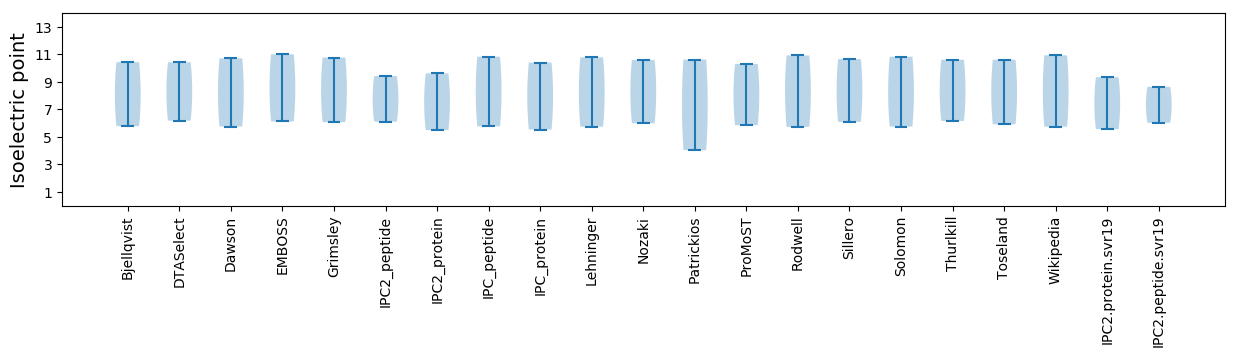
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE98|A0A1L3KE98_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 10 OX=1922681 PE=4 SV=1
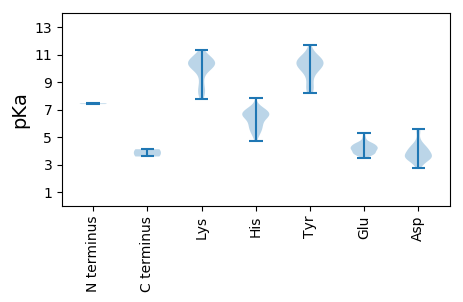
MM1 pKa = 7.45ISSIAEE7 pKa = 3.71FLGNQLLLPFDD18 pKa = 3.9KK19 pKa = 11.17VVGLVAEE26 pKa = 4.42KK27 pKa = 10.93GKK29 pKa = 10.09LQVPTRR35 pKa = 11.84EE36 pKa = 4.05SFLGYY41 pKa = 10.54VVMAALTLAIAEE53 pKa = 4.53FFRR56 pKa = 11.84AHH58 pKa = 6.65GNLCGRR64 pKa = 11.84AYY66 pKa = 10.76GHH68 pKa = 6.67FLGACASLWKK78 pKa = 10.49YY79 pKa = 10.64CVDD82 pKa = 3.34RR83 pKa = 11.84MQRR86 pKa = 11.84PATGDD91 pKa = 3.34VASQRR96 pKa = 11.84FGYY99 pKa = 8.81FQHH102 pKa = 7.05GDD104 pKa = 2.91EE105 pKa = 5.0HH106 pKa = 6.48YY107 pKa = 8.35WRR109 pKa = 11.84CRR111 pKa = 11.84RR112 pKa = 11.84TSKK115 pKa = 10.66LYY117 pKa = 10.01RR118 pKa = 11.84VEE120 pKa = 4.05PGRR123 pKa = 11.84GQFYY127 pKa = 10.36IDD129 pKa = 3.67DD130 pKa = 4.6AGVEE134 pKa = 4.32QYY136 pKa = 11.41EE137 pKa = 4.44VMDD140 pKa = 4.4YY141 pKa = 11.06IHH143 pKa = 6.73VLVQGSEE150 pKa = 4.39LIQPEE155 pKa = 4.13ANLLHH160 pKa = 6.85GEE162 pKa = 4.21VPLPFEE168 pKa = 4.65GLHH171 pKa = 6.06KK172 pKa = 10.7SVVAIFQKK180 pKa = 10.43NHH182 pKa = 6.69DD183 pKa = 4.05GEE185 pKa = 4.56FAQVGHH191 pKa = 5.84GHH193 pKa = 6.64RR194 pKa = 11.84CGDD197 pKa = 3.55YY198 pKa = 10.85LFTNAHH204 pKa = 4.93VHH206 pKa = 6.58DD207 pKa = 4.37ATNGDD212 pKa = 3.98VYY214 pKa = 10.81FSKK217 pKa = 10.57DD218 pKa = 3.39LKK220 pKa = 10.95KK221 pKa = 10.11FWPVGEE227 pKa = 3.73FTYY230 pKa = 10.16FVKK233 pKa = 10.67NYY235 pKa = 10.52SKK237 pKa = 10.62ATGNDD242 pKa = 2.71LGAYY246 pKa = 8.2FVSRR250 pKa = 11.84GTWAASGVSTFQATNYY266 pKa = 8.23WPQSEE271 pKa = 4.04GRR273 pKa = 11.84IEE275 pKa = 3.57ITAYY279 pKa = 10.43DD280 pKa = 4.08DD281 pKa = 3.62VRR283 pKa = 11.84GKK285 pKa = 10.34HH286 pKa = 5.91MIAKK290 pKa = 8.69GHH292 pKa = 6.3VYY294 pKa = 10.51NQTLVDD300 pKa = 4.05AKK302 pKa = 10.82LGMVPHH308 pKa = 6.36SAITKK313 pKa = 8.36VGWSGAPVFMMKK325 pKa = 10.0GQKK328 pKa = 8.48RR329 pKa = 11.84TIVGIHH335 pKa = 6.66CGGGSDD341 pKa = 4.31GSANYY346 pKa = 10.04ACSIYY351 pKa = 10.68EE352 pKa = 3.7LLEE355 pKa = 3.57FRR357 pKa = 11.84RR358 pKa = 11.84RR359 pKa = 11.84LGLEE363 pKa = 3.46PKK365 pKa = 10.18IPLVCEE371 pKa = 4.17AKK373 pKa = 10.61FISPPSKK380 pKa = 8.93TKK382 pKa = 10.56QFDD385 pKa = 3.7RR386 pKa = 11.84YY387 pKa = 10.57DD388 pKa = 3.59EE389 pKa = 4.39EE390 pKa = 4.9SPEE393 pKa = 3.81EE394 pKa = 3.93RR395 pKa = 11.84EE396 pKa = 3.9RR397 pKa = 11.84RR398 pKa = 11.84EE399 pKa = 3.85DD400 pKa = 3.63SEE402 pKa = 4.19MLTRR406 pKa = 11.84EE407 pKa = 4.23AEE409 pKa = 4.12EE410 pKa = 4.32HH411 pKa = 4.68VTEE414 pKa = 4.61LLHH417 pKa = 7.81GEE419 pKa = 4.23ANSPPKK425 pKa = 10.22KK426 pKa = 10.34DD427 pKa = 3.42ISPEE431 pKa = 3.81AKK433 pKa = 10.4LPGPTWYY440 pKa = 9.61EE441 pKa = 3.8HH442 pKa = 5.79CHH444 pKa = 5.68IDD446 pKa = 4.93APHH449 pKa = 6.42SPAGTLVLHH458 pKa = 6.95LADD461 pKa = 5.29DD462 pKa = 4.22VANMADD468 pKa = 5.61LEE470 pKa = 4.65DD471 pKa = 5.58LDD473 pKa = 5.45IPDD476 pKa = 5.37GPPSSPDD483 pKa = 3.4PDD485 pKa = 4.39LDD487 pKa = 4.24GGAADD492 pKa = 3.48VHH494 pKa = 6.58VNAVLAEE501 pKa = 4.15YY502 pKa = 10.26ILRR505 pKa = 11.84ARR507 pKa = 11.84AINEE511 pKa = 4.18EE512 pKa = 4.28VGPPPGLFPSLSEE525 pKa = 4.17AEE527 pKa = 4.22PPEE530 pKa = 4.47VPQEE534 pKa = 3.8EE535 pKa = 4.9HH536 pKa = 6.59EE537 pKa = 4.35PEE539 pKa = 3.99EE540 pKa = 4.52HH541 pKa = 6.78GGLSEE546 pKa = 5.02PIYY549 pKa = 10.02MGNPQGEE556 pKa = 4.59PVTLGPQISPEE567 pKa = 3.81EE568 pKa = 4.11RR569 pKa = 11.84EE570 pKa = 4.03RR571 pKa = 11.84GQKK574 pKa = 8.41TFEE577 pKa = 4.22SRR579 pKa = 11.84ASRR582 pKa = 11.84PLWAKK587 pKa = 10.41AFLGVGLLGATALSPSGRR605 pKa = 11.84AVTDD609 pKa = 3.56EE610 pKa = 4.25KK611 pKa = 11.31ARR613 pKa = 11.84SLLRR617 pKa = 11.84CSPQDD622 pKa = 3.32LAEE625 pKa = 4.16ARR627 pKa = 11.84KK628 pKa = 8.24MCPISLLATPAFDD641 pKa = 4.35RR642 pKa = 11.84YY643 pKa = 10.13RR644 pKa = 11.84GYY646 pKa = 10.83LGSGDD651 pKa = 3.71VDD653 pKa = 3.31WVNFEE658 pKa = 3.77SHH660 pKa = 7.04KK661 pKa = 10.59IVNQSNVTVAATVGTSRR678 pKa = 11.84LYY680 pKa = 11.03SKK682 pKa = 7.76EE683 pKa = 4.15TPLTKK688 pKa = 10.61LPGRR692 pKa = 11.84YY693 pKa = 8.53RR694 pKa = 11.84EE695 pKa = 4.52VISEE699 pKa = 4.06MGLGPEE705 pKa = 4.47QGFEE709 pKa = 4.45DD710 pKa = 4.64YY711 pKa = 11.7VMPPSGAEE719 pKa = 3.69AVKK722 pKa = 10.57EE723 pKa = 4.11SLEE726 pKa = 4.2SQLNATCHH734 pKa = 5.49EE735 pKa = 4.29PWPKK739 pKa = 10.37EE740 pKa = 3.66VLEE743 pKa = 4.4RR744 pKa = 11.84DD745 pKa = 3.12GGNYY749 pKa = 9.92LRR751 pKa = 11.84DD752 pKa = 3.88EE753 pKa = 4.41LHH755 pKa = 6.69SEE757 pKa = 3.8AGKK760 pKa = 10.93YY761 pKa = 9.38PLAFSSGISQVHH773 pKa = 6.24DD774 pKa = 3.55FVVKK778 pKa = 10.31LAKK781 pKa = 10.5EE782 pKa = 3.85FDD784 pKa = 3.91GTKK787 pKa = 10.07SSGWSAHH794 pKa = 5.17YY795 pKa = 10.52RR796 pKa = 11.84PGPKK800 pKa = 9.74SAWQCAEE807 pKa = 3.7GLEE810 pKa = 4.38TASYY814 pKa = 10.73LVRR817 pKa = 11.84CRR819 pKa = 11.84LLLRR823 pKa = 11.84LAWGAHH829 pKa = 5.35EE830 pKa = 4.26MSKK833 pKa = 7.89MTPEE837 pKa = 3.75QMVQNGLSDD846 pKa = 3.88PKK848 pKa = 10.37VASIKK853 pKa = 10.41SEE855 pKa = 3.52PHH857 pKa = 6.05APRR860 pKa = 11.84KK861 pKa = 9.25AANKK865 pKa = 7.88KK866 pKa = 8.24WRR868 pKa = 11.84VIWGASIIDD877 pKa = 4.2LLAQGVTCRR886 pKa = 11.84IQDD889 pKa = 3.39KK890 pKa = 11.03LDD892 pKa = 2.95IHH894 pKa = 7.02SYY896 pKa = 10.4QDD898 pKa = 3.02GTGQHH903 pKa = 5.57SQAVGLGHH911 pKa = 6.65HH912 pKa = 7.2PEE914 pKa = 5.29GIQMIGKK921 pKa = 9.25HH922 pKa = 5.26IEE924 pKa = 3.74RR925 pKa = 11.84LQATGHH931 pKa = 5.43QLFDD935 pKa = 4.16ADD937 pKa = 3.61ASGWDD942 pKa = 3.43MSVKK946 pKa = 9.96RR947 pKa = 11.84DD948 pKa = 4.03SILMDD953 pKa = 3.29AEE955 pKa = 3.94RR956 pKa = 11.84RR957 pKa = 11.84IRR959 pKa = 11.84CYY961 pKa = 9.96VGPYY965 pKa = 9.97HH966 pKa = 7.17EE967 pKa = 5.32IFAALQMAEE976 pKa = 3.92AMTNSAHH983 pKa = 5.91MLAFGPYY990 pKa = 9.9LFSIYY995 pKa = 10.23LAGITASGILPTTSQNSFMRR1015 pKa = 11.84AFIARR1020 pKa = 11.84LVGIIATLCGGDD1032 pKa = 4.03DD1033 pKa = 3.92LMGAGKK1039 pKa = 10.07FDD1041 pKa = 3.77PAYY1044 pKa = 9.59EE1045 pKa = 4.03RR1046 pKa = 11.84RR1047 pKa = 11.84FGPIEE1052 pKa = 3.85KK1053 pKa = 10.02KK1054 pKa = 8.34ITYY1057 pKa = 9.39RR1058 pKa = 11.84EE1059 pKa = 3.7QGEE1062 pKa = 4.39PIEE1065 pKa = 4.19FTSFLFRR1072 pKa = 11.84KK1073 pKa = 9.7EE1074 pKa = 3.8NGVWKK1079 pKa = 10.57ADD1081 pKa = 3.55FVNLGKK1087 pKa = 10.28SCAKK1091 pKa = 10.18LAFSSKK1097 pKa = 10.22PVTPDD1102 pKa = 2.81QLGGVLYY1109 pKa = 10.37HH1110 pKa = 7.05LRR1112 pKa = 11.84DD1113 pKa = 3.74NEE1115 pKa = 4.59SNASLFRR1122 pKa = 11.84EE1123 pKa = 4.68ICRR1126 pKa = 11.84KK1127 pKa = 8.8MDD1129 pKa = 3.21WPIEE1133 pKa = 4.0GAIAGPDD1140 pKa = 3.56GDD1142 pKa = 3.95QGG1144 pKa = 3.58
MM1 pKa = 7.45ISSIAEE7 pKa = 3.71FLGNQLLLPFDD18 pKa = 3.9KK19 pKa = 11.17VVGLVAEE26 pKa = 4.42KK27 pKa = 10.93GKK29 pKa = 10.09LQVPTRR35 pKa = 11.84EE36 pKa = 4.05SFLGYY41 pKa = 10.54VVMAALTLAIAEE53 pKa = 4.53FFRR56 pKa = 11.84AHH58 pKa = 6.65GNLCGRR64 pKa = 11.84AYY66 pKa = 10.76GHH68 pKa = 6.67FLGACASLWKK78 pKa = 10.49YY79 pKa = 10.64CVDD82 pKa = 3.34RR83 pKa = 11.84MQRR86 pKa = 11.84PATGDD91 pKa = 3.34VASQRR96 pKa = 11.84FGYY99 pKa = 8.81FQHH102 pKa = 7.05GDD104 pKa = 2.91EE105 pKa = 5.0HH106 pKa = 6.48YY107 pKa = 8.35WRR109 pKa = 11.84CRR111 pKa = 11.84RR112 pKa = 11.84TSKK115 pKa = 10.66LYY117 pKa = 10.01RR118 pKa = 11.84VEE120 pKa = 4.05PGRR123 pKa = 11.84GQFYY127 pKa = 10.36IDD129 pKa = 3.67DD130 pKa = 4.6AGVEE134 pKa = 4.32QYY136 pKa = 11.41EE137 pKa = 4.44VMDD140 pKa = 4.4YY141 pKa = 11.06IHH143 pKa = 6.73VLVQGSEE150 pKa = 4.39LIQPEE155 pKa = 4.13ANLLHH160 pKa = 6.85GEE162 pKa = 4.21VPLPFEE168 pKa = 4.65GLHH171 pKa = 6.06KK172 pKa = 10.7SVVAIFQKK180 pKa = 10.43NHH182 pKa = 6.69DD183 pKa = 4.05GEE185 pKa = 4.56FAQVGHH191 pKa = 5.84GHH193 pKa = 6.64RR194 pKa = 11.84CGDD197 pKa = 3.55YY198 pKa = 10.85LFTNAHH204 pKa = 4.93VHH206 pKa = 6.58DD207 pKa = 4.37ATNGDD212 pKa = 3.98VYY214 pKa = 10.81FSKK217 pKa = 10.57DD218 pKa = 3.39LKK220 pKa = 10.95KK221 pKa = 10.11FWPVGEE227 pKa = 3.73FTYY230 pKa = 10.16FVKK233 pKa = 10.67NYY235 pKa = 10.52SKK237 pKa = 10.62ATGNDD242 pKa = 2.71LGAYY246 pKa = 8.2FVSRR250 pKa = 11.84GTWAASGVSTFQATNYY266 pKa = 8.23WPQSEE271 pKa = 4.04GRR273 pKa = 11.84IEE275 pKa = 3.57ITAYY279 pKa = 10.43DD280 pKa = 4.08DD281 pKa = 3.62VRR283 pKa = 11.84GKK285 pKa = 10.34HH286 pKa = 5.91MIAKK290 pKa = 8.69GHH292 pKa = 6.3VYY294 pKa = 10.51NQTLVDD300 pKa = 4.05AKK302 pKa = 10.82LGMVPHH308 pKa = 6.36SAITKK313 pKa = 8.36VGWSGAPVFMMKK325 pKa = 10.0GQKK328 pKa = 8.48RR329 pKa = 11.84TIVGIHH335 pKa = 6.66CGGGSDD341 pKa = 4.31GSANYY346 pKa = 10.04ACSIYY351 pKa = 10.68EE352 pKa = 3.7LLEE355 pKa = 3.57FRR357 pKa = 11.84RR358 pKa = 11.84RR359 pKa = 11.84LGLEE363 pKa = 3.46PKK365 pKa = 10.18IPLVCEE371 pKa = 4.17AKK373 pKa = 10.61FISPPSKK380 pKa = 8.93TKK382 pKa = 10.56QFDD385 pKa = 3.7RR386 pKa = 11.84YY387 pKa = 10.57DD388 pKa = 3.59EE389 pKa = 4.39EE390 pKa = 4.9SPEE393 pKa = 3.81EE394 pKa = 3.93RR395 pKa = 11.84EE396 pKa = 3.9RR397 pKa = 11.84RR398 pKa = 11.84EE399 pKa = 3.85DD400 pKa = 3.63SEE402 pKa = 4.19MLTRR406 pKa = 11.84EE407 pKa = 4.23AEE409 pKa = 4.12EE410 pKa = 4.32HH411 pKa = 4.68VTEE414 pKa = 4.61LLHH417 pKa = 7.81GEE419 pKa = 4.23ANSPPKK425 pKa = 10.22KK426 pKa = 10.34DD427 pKa = 3.42ISPEE431 pKa = 3.81AKK433 pKa = 10.4LPGPTWYY440 pKa = 9.61EE441 pKa = 3.8HH442 pKa = 5.79CHH444 pKa = 5.68IDD446 pKa = 4.93APHH449 pKa = 6.42SPAGTLVLHH458 pKa = 6.95LADD461 pKa = 5.29DD462 pKa = 4.22VANMADD468 pKa = 5.61LEE470 pKa = 4.65DD471 pKa = 5.58LDD473 pKa = 5.45IPDD476 pKa = 5.37GPPSSPDD483 pKa = 3.4PDD485 pKa = 4.39LDD487 pKa = 4.24GGAADD492 pKa = 3.48VHH494 pKa = 6.58VNAVLAEE501 pKa = 4.15YY502 pKa = 10.26ILRR505 pKa = 11.84ARR507 pKa = 11.84AINEE511 pKa = 4.18EE512 pKa = 4.28VGPPPGLFPSLSEE525 pKa = 4.17AEE527 pKa = 4.22PPEE530 pKa = 4.47VPQEE534 pKa = 3.8EE535 pKa = 4.9HH536 pKa = 6.59EE537 pKa = 4.35PEE539 pKa = 3.99EE540 pKa = 4.52HH541 pKa = 6.78GGLSEE546 pKa = 5.02PIYY549 pKa = 10.02MGNPQGEE556 pKa = 4.59PVTLGPQISPEE567 pKa = 3.81EE568 pKa = 4.11RR569 pKa = 11.84EE570 pKa = 4.03RR571 pKa = 11.84GQKK574 pKa = 8.41TFEE577 pKa = 4.22SRR579 pKa = 11.84ASRR582 pKa = 11.84PLWAKK587 pKa = 10.41AFLGVGLLGATALSPSGRR605 pKa = 11.84AVTDD609 pKa = 3.56EE610 pKa = 4.25KK611 pKa = 11.31ARR613 pKa = 11.84SLLRR617 pKa = 11.84CSPQDD622 pKa = 3.32LAEE625 pKa = 4.16ARR627 pKa = 11.84KK628 pKa = 8.24MCPISLLATPAFDD641 pKa = 4.35RR642 pKa = 11.84YY643 pKa = 10.13RR644 pKa = 11.84GYY646 pKa = 10.83LGSGDD651 pKa = 3.71VDD653 pKa = 3.31WVNFEE658 pKa = 3.77SHH660 pKa = 7.04KK661 pKa = 10.59IVNQSNVTVAATVGTSRR678 pKa = 11.84LYY680 pKa = 11.03SKK682 pKa = 7.76EE683 pKa = 4.15TPLTKK688 pKa = 10.61LPGRR692 pKa = 11.84YY693 pKa = 8.53RR694 pKa = 11.84EE695 pKa = 4.52VISEE699 pKa = 4.06MGLGPEE705 pKa = 4.47QGFEE709 pKa = 4.45DD710 pKa = 4.64YY711 pKa = 11.7VMPPSGAEE719 pKa = 3.69AVKK722 pKa = 10.57EE723 pKa = 4.11SLEE726 pKa = 4.2SQLNATCHH734 pKa = 5.49EE735 pKa = 4.29PWPKK739 pKa = 10.37EE740 pKa = 3.66VLEE743 pKa = 4.4RR744 pKa = 11.84DD745 pKa = 3.12GGNYY749 pKa = 9.92LRR751 pKa = 11.84DD752 pKa = 3.88EE753 pKa = 4.41LHH755 pKa = 6.69SEE757 pKa = 3.8AGKK760 pKa = 10.93YY761 pKa = 9.38PLAFSSGISQVHH773 pKa = 6.24DD774 pKa = 3.55FVVKK778 pKa = 10.31LAKK781 pKa = 10.5EE782 pKa = 3.85FDD784 pKa = 3.91GTKK787 pKa = 10.07SSGWSAHH794 pKa = 5.17YY795 pKa = 10.52RR796 pKa = 11.84PGPKK800 pKa = 9.74SAWQCAEE807 pKa = 3.7GLEE810 pKa = 4.38TASYY814 pKa = 10.73LVRR817 pKa = 11.84CRR819 pKa = 11.84LLLRR823 pKa = 11.84LAWGAHH829 pKa = 5.35EE830 pKa = 4.26MSKK833 pKa = 7.89MTPEE837 pKa = 3.75QMVQNGLSDD846 pKa = 3.88PKK848 pKa = 10.37VASIKK853 pKa = 10.41SEE855 pKa = 3.52PHH857 pKa = 6.05APRR860 pKa = 11.84KK861 pKa = 9.25AANKK865 pKa = 7.88KK866 pKa = 8.24WRR868 pKa = 11.84VIWGASIIDD877 pKa = 4.2LLAQGVTCRR886 pKa = 11.84IQDD889 pKa = 3.39KK890 pKa = 11.03LDD892 pKa = 2.95IHH894 pKa = 7.02SYY896 pKa = 10.4QDD898 pKa = 3.02GTGQHH903 pKa = 5.57SQAVGLGHH911 pKa = 6.65HH912 pKa = 7.2PEE914 pKa = 5.29GIQMIGKK921 pKa = 9.25HH922 pKa = 5.26IEE924 pKa = 3.74RR925 pKa = 11.84LQATGHH931 pKa = 5.43QLFDD935 pKa = 4.16ADD937 pKa = 3.61ASGWDD942 pKa = 3.43MSVKK946 pKa = 9.96RR947 pKa = 11.84DD948 pKa = 4.03SILMDD953 pKa = 3.29AEE955 pKa = 3.94RR956 pKa = 11.84RR957 pKa = 11.84IRR959 pKa = 11.84CYY961 pKa = 9.96VGPYY965 pKa = 9.97HH966 pKa = 7.17EE967 pKa = 5.32IFAALQMAEE976 pKa = 3.92AMTNSAHH983 pKa = 5.91MLAFGPYY990 pKa = 9.9LFSIYY995 pKa = 10.23LAGITASGILPTTSQNSFMRR1015 pKa = 11.84AFIARR1020 pKa = 11.84LVGIIATLCGGDD1032 pKa = 4.03DD1033 pKa = 3.92LMGAGKK1039 pKa = 10.07FDD1041 pKa = 3.77PAYY1044 pKa = 9.59EE1045 pKa = 4.03RR1046 pKa = 11.84RR1047 pKa = 11.84FGPIEE1052 pKa = 3.85KK1053 pKa = 10.02KK1054 pKa = 8.34ITYY1057 pKa = 9.39RR1058 pKa = 11.84EE1059 pKa = 3.7QGEE1062 pKa = 4.39PIEE1065 pKa = 4.19FTSFLFRR1072 pKa = 11.84KK1073 pKa = 9.7EE1074 pKa = 3.8NGVWKK1079 pKa = 10.57ADD1081 pKa = 3.55FVNLGKK1087 pKa = 10.28SCAKK1091 pKa = 10.18LAFSSKK1097 pKa = 10.22PVTPDD1102 pKa = 2.81QLGGVLYY1109 pKa = 10.37HH1110 pKa = 7.05LRR1112 pKa = 11.84DD1113 pKa = 3.74NEE1115 pKa = 4.59SNASLFRR1122 pKa = 11.84EE1123 pKa = 4.68ICRR1126 pKa = 11.84KK1127 pKa = 8.8MDD1129 pKa = 3.21WPIEE1133 pKa = 4.0GAIAGPDD1140 pKa = 3.56GDD1142 pKa = 3.95QGG1144 pKa = 3.58
Molecular weight: 126.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE98|A0A1L3KE98_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 10 OX=1922681 PE=4 SV=1
MM1 pKa = 7.4SSKK4 pKa = 10.26GRR6 pKa = 11.84QARR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84NRR13 pKa = 11.84RR14 pKa = 11.84AAAQPRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84HH23 pKa = 5.26LKK25 pKa = 9.98GPGGGAMSRR34 pKa = 11.84GSRR37 pKa = 11.84PTNRR41 pKa = 11.84SHH43 pKa = 6.31ATNVLATGVAAAPKK57 pKa = 9.88KK58 pKa = 10.9AFGCTIGHH66 pKa = 6.46TLHH69 pKa = 6.82CWDD72 pKa = 4.72AKK74 pKa = 10.61HH75 pKa = 6.05PHH77 pKa = 6.69HH78 pKa = 7.02LPLPRR83 pKa = 11.84AVGPYY88 pKa = 8.84TVIRR92 pKa = 11.84ATKK95 pKa = 9.93RR96 pKa = 11.84ITSGRR101 pKa = 11.84AANIIGSFQFKK112 pKa = 10.27DD113 pKa = 3.95GASTTPDD120 pKa = 3.07FGNWSEE126 pKa = 4.46TIMVSDD132 pKa = 4.53VNPTAPINQSNNTFASSVEE151 pKa = 3.99LGGLGAAATLVPSALSVQIMCPTALQTASGVIYY184 pKa = 10.29AGVMNTQAALAGRR197 pKa = 11.84TEE199 pKa = 3.95TWFSYY204 pKa = 8.96MEE206 pKa = 4.52KK207 pKa = 9.94FVQFQSPRR215 pKa = 11.84LLAASKK221 pKa = 10.15LALRR225 pKa = 11.84GVQINSYY232 pKa = 8.5PLNMSEE238 pKa = 4.37VSKK241 pKa = 9.27FTQLKK246 pKa = 8.17GTGDD250 pKa = 3.59EE251 pKa = 4.14NFAWEE256 pKa = 4.5HH257 pKa = 5.77FNPAGWAPIVIYY269 pKa = 10.55NPSGTTLEE277 pKa = 3.9MLITVEE283 pKa = 3.67YY284 pKa = 10.05RR285 pKa = 11.84VRR287 pKa = 11.84FDD289 pKa = 4.28LDD291 pKa = 3.4HH292 pKa = 7.03PASASHH298 pKa = 5.65SHH300 pKa = 6.47HH301 pKa = 6.59PVASDD306 pKa = 3.64SAWDD310 pKa = 3.61RR311 pKa = 11.84LVRR314 pKa = 11.84QASSLGNGVMDD325 pKa = 4.13IADD328 pKa = 3.63VVANAGMSVGRR339 pKa = 11.84AIAVANRR346 pKa = 11.84VNNVLRR352 pKa = 11.84PLPLAAAAAA361 pKa = 4.12
MM1 pKa = 7.4SSKK4 pKa = 10.26GRR6 pKa = 11.84QARR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84NRR13 pKa = 11.84RR14 pKa = 11.84AAAQPRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84HH23 pKa = 5.26LKK25 pKa = 9.98GPGGGAMSRR34 pKa = 11.84GSRR37 pKa = 11.84PTNRR41 pKa = 11.84SHH43 pKa = 6.31ATNVLATGVAAAPKK57 pKa = 9.88KK58 pKa = 10.9AFGCTIGHH66 pKa = 6.46TLHH69 pKa = 6.82CWDD72 pKa = 4.72AKK74 pKa = 10.61HH75 pKa = 6.05PHH77 pKa = 6.69HH78 pKa = 7.02LPLPRR83 pKa = 11.84AVGPYY88 pKa = 8.84TVIRR92 pKa = 11.84ATKK95 pKa = 9.93RR96 pKa = 11.84ITSGRR101 pKa = 11.84AANIIGSFQFKK112 pKa = 10.27DD113 pKa = 3.95GASTTPDD120 pKa = 3.07FGNWSEE126 pKa = 4.46TIMVSDD132 pKa = 4.53VNPTAPINQSNNTFASSVEE151 pKa = 3.99LGGLGAAATLVPSALSVQIMCPTALQTASGVIYY184 pKa = 10.29AGVMNTQAALAGRR197 pKa = 11.84TEE199 pKa = 3.95TWFSYY204 pKa = 8.96MEE206 pKa = 4.52KK207 pKa = 9.94FVQFQSPRR215 pKa = 11.84LLAASKK221 pKa = 10.15LALRR225 pKa = 11.84GVQINSYY232 pKa = 8.5PLNMSEE238 pKa = 4.37VSKK241 pKa = 9.27FTQLKK246 pKa = 8.17GTGDD250 pKa = 3.59EE251 pKa = 4.14NFAWEE256 pKa = 4.5HH257 pKa = 5.77FNPAGWAPIVIYY269 pKa = 10.55NPSGTTLEE277 pKa = 3.9MLITVEE283 pKa = 3.67YY284 pKa = 10.05RR285 pKa = 11.84VRR287 pKa = 11.84FDD289 pKa = 4.28LDD291 pKa = 3.4HH292 pKa = 7.03PASASHH298 pKa = 5.65SHH300 pKa = 6.47HH301 pKa = 6.59PVASDD306 pKa = 3.64SAWDD310 pKa = 3.61RR311 pKa = 11.84LVRR314 pKa = 11.84QASSLGNGVMDD325 pKa = 4.13IADD328 pKa = 3.63VVANAGMSVGRR339 pKa = 11.84AIAVANRR346 pKa = 11.84VNNVLRR352 pKa = 11.84PLPLAAAAAA361 pKa = 4.12
Molecular weight: 38.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1505 |
361 |
1144 |
752.5 |
82.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.1 ± 2.014 | 1.462 ± 0.315 |
4.784 ± 0.868 | 6.445 ± 1.976 |
3.854 ± 0.265 | 8.97 ± 0.468 |
3.522 ± 0.099 | 4.385 ± 0.115 |
4.518 ± 0.736 | 8.04 ± 0.557 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.392 ± 0.189 | 3.256 ± 1.142 |
6.179 ± 0.043 | 3.522 ± 0.099 |
5.847 ± 0.677 | 7.243 ± 0.672 |
4.585 ± 1.032 | 6.246 ± 0.478 |
1.595 ± 0.034 | 3.056 ± 0.697 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
