
Shahe narna-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.13
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KII2|A0A1L3KII2_9VIRU RNA-dependent RNA polymerase OS=Shahe narna-like virus 3 OX=1923431 PE=4 SV=1
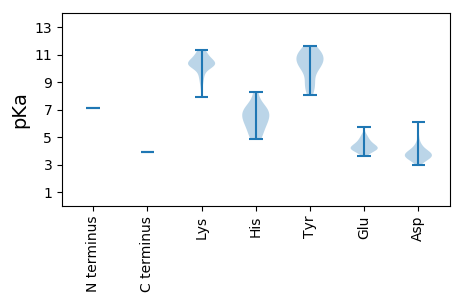
MM1 pKa = 7.12FLKK4 pKa = 10.52RR5 pKa = 11.84SLAPVPRR12 pKa = 11.84SLIKK16 pKa = 10.4KK17 pKa = 9.94SRR19 pKa = 11.84DD20 pKa = 3.36DD21 pKa = 3.73HH22 pKa = 6.9KK23 pKa = 11.22STLTTPSEE31 pKa = 4.14RR32 pKa = 11.84VHH34 pKa = 7.26DD35 pKa = 3.98VTLKK39 pKa = 10.91FIEE42 pKa = 4.91DD43 pKa = 3.85CTTVYY48 pKa = 9.84TRR50 pKa = 11.84SLTKK54 pKa = 10.23CRR56 pKa = 11.84QEE58 pKa = 4.28RR59 pKa = 11.84LEE61 pKa = 4.67KK62 pKa = 10.27PAISPINQLKK72 pKa = 9.89RR73 pKa = 11.84LKK75 pKa = 9.34RR76 pKa = 11.84TYY78 pKa = 10.96GPIEE82 pKa = 4.06VKK84 pKa = 10.21EE85 pKa = 4.22DD86 pKa = 3.64CLISCEE92 pKa = 4.51DD93 pKa = 3.81EE94 pKa = 4.15SSHH97 pKa = 6.68RR98 pKa = 11.84QLDD101 pKa = 3.94DD102 pKa = 3.67SSLSCSEE109 pKa = 4.25TPFLNLMATKK119 pKa = 9.74EE120 pKa = 3.96LHH122 pKa = 4.86NTRR125 pKa = 11.84DD126 pKa = 3.48RR127 pKa = 11.84NQVHH131 pKa = 6.54GLHH134 pKa = 6.09SLNRR138 pKa = 11.84SFYY141 pKa = 8.8PLSYY145 pKa = 10.12KK146 pKa = 10.21IKK148 pKa = 10.65EE149 pKa = 3.79VDD151 pKa = 3.32QNLNTFDD158 pKa = 4.34NCWIDD163 pKa = 3.56QKK165 pKa = 11.35VKK167 pKa = 10.33EE168 pKa = 4.09SSYY171 pKa = 10.58YY172 pKa = 10.88DD173 pKa = 3.51KK174 pKa = 11.29LKK176 pKa = 10.99SDD178 pKa = 3.66TKK180 pKa = 10.14LTEE183 pKa = 3.88EE184 pKa = 5.15GEE186 pKa = 4.66FISQQDD192 pKa = 2.94ISMSKK197 pKa = 10.72YY198 pKa = 10.97SLDD201 pKa = 4.6KK202 pKa = 10.75INKK205 pKa = 8.76IYY207 pKa = 10.12PLSKK211 pKa = 9.92HH212 pKa = 5.91SSYY215 pKa = 11.58NNSVKK220 pKa = 10.43IGGAQSEE227 pKa = 4.45ISLNLQNSNLKK238 pKa = 10.46LGSNNGGLRR247 pKa = 11.84KK248 pKa = 9.02FAMLNNAMKK257 pKa = 10.34NQYY260 pKa = 9.8EE261 pKa = 4.09IAKK264 pKa = 9.53AKK266 pKa = 10.09CLNEE270 pKa = 3.95SKK272 pKa = 10.76DD273 pKa = 3.51IIKK276 pKa = 10.54EE277 pKa = 4.14SVRR280 pKa = 11.84VCSIVEE286 pKa = 3.84PLKK289 pKa = 10.93VRR291 pKa = 11.84TVTAEE296 pKa = 3.6EE297 pKa = 4.39ARR299 pKa = 11.84NQFIKK304 pKa = 10.59PIQKK308 pKa = 10.25LLWEE312 pKa = 5.12DD313 pKa = 3.88LLSHH317 pKa = 7.04EE318 pKa = 5.72DD319 pKa = 4.02FFLTRR324 pKa = 11.84NEE326 pKa = 4.26DD327 pKa = 5.06FEE329 pKa = 5.18SHH331 pKa = 5.4WNRR334 pKa = 11.84KK335 pKa = 7.92FKK337 pKa = 10.88FCDD340 pKa = 3.38LPYY343 pKa = 10.35HH344 pKa = 7.03ISGDD348 pKa = 3.81YY349 pKa = 10.19KK350 pKa = 11.14AATDD354 pKa = 3.66NLNRR358 pKa = 11.84NVINVVITQLSTLLPTNLIDD378 pKa = 6.1LFMKK382 pKa = 10.66NAGLHH387 pKa = 5.02YY388 pKa = 10.59LDD390 pKa = 4.29YY391 pKa = 11.62NEE393 pKa = 4.43VAFSLRR399 pKa = 11.84EE400 pKa = 4.07SPDD403 pKa = 3.47KK404 pKa = 11.18IGDD407 pKa = 3.52VYY409 pKa = 10.88LQKK412 pKa = 10.74NGQLMGSLTSFPLLCFINWISYY434 pKa = 8.03QFSKK438 pKa = 11.23YY439 pKa = 10.27LVNNYY444 pKa = 9.29PDD446 pKa = 3.34EE447 pKa = 4.59HH448 pKa = 8.31LSEE451 pKa = 4.48SCAINGDD458 pKa = 3.98DD459 pKa = 4.46IYY461 pKa = 11.4FKK463 pKa = 11.23ASLKK467 pKa = 10.71GYY469 pKa = 10.23AKK471 pKa = 8.86WQEE474 pKa = 4.17VVHH477 pKa = 6.4SFGLSPSPGKK487 pKa = 10.38NYY489 pKa = 10.39SSSNCFLINSRR500 pKa = 11.84NFLVNTKK507 pKa = 10.21SEE509 pKa = 4.18LKK511 pKa = 10.01EE512 pKa = 3.64IPFINFQMLPQYY524 pKa = 11.06SEE526 pKa = 4.15DD527 pKa = 3.67SNNKK531 pKa = 9.17FEE533 pKa = 4.84HH534 pKa = 5.93NKK536 pKa = 10.2KK537 pKa = 10.09EE538 pKa = 4.81LLNQMKK544 pKa = 10.17GDD546 pKa = 3.79SVYY549 pKa = 11.2SEE551 pKa = 4.42VNSPIGIIFRR561 pKa = 11.84TFCKK565 pKa = 9.67QANINLVKK573 pKa = 10.2PSKK576 pKa = 10.62FDD578 pKa = 3.1IQLIYY583 pKa = 10.86YY584 pKa = 8.43FYY586 pKa = 11.15SVWKK590 pKa = 10.37KK591 pKa = 10.66EE592 pKa = 3.98IIKK595 pKa = 10.28CPRR598 pKa = 11.84EE599 pKa = 3.94FYY601 pKa = 10.85LPSLIGGMGALTLDD615 pKa = 3.77EE616 pKa = 4.6FKK618 pKa = 10.27KK619 pKa = 9.61TNLLTKK625 pKa = 9.25TKK627 pKa = 10.23KK628 pKa = 10.48LSLLRR633 pKa = 11.84GFKK636 pKa = 10.39CKK638 pKa = 10.43SCFYY642 pKa = 8.95MQTYY646 pKa = 8.39EE647 pKa = 4.92QIGSDD652 pKa = 3.31KK653 pKa = 11.25DD654 pKa = 3.66QIEE657 pKa = 4.34SFAAKK662 pKa = 10.14LEE664 pKa = 4.15NIILDD669 pKa = 4.15EE670 pKa = 4.75LSGTDD675 pKa = 4.01PEE677 pKa = 4.33NKK679 pKa = 9.52QLSSKK684 pKa = 10.82SLLSPVSTFVSKK696 pKa = 11.12GGVEE700 pKa = 4.4KK701 pKa = 10.45YY702 pKa = 8.96TLQKK706 pKa = 10.53FFKK709 pKa = 10.18KK710 pKa = 10.16LGKK713 pKa = 10.12SSFRR717 pKa = 11.84NRR719 pKa = 11.84KK720 pKa = 8.4FKK722 pKa = 10.34PATVLSIEE730 pKa = 4.22KK731 pKa = 10.6SLL733 pKa = 3.9
MM1 pKa = 7.12FLKK4 pKa = 10.52RR5 pKa = 11.84SLAPVPRR12 pKa = 11.84SLIKK16 pKa = 10.4KK17 pKa = 9.94SRR19 pKa = 11.84DD20 pKa = 3.36DD21 pKa = 3.73HH22 pKa = 6.9KK23 pKa = 11.22STLTTPSEE31 pKa = 4.14RR32 pKa = 11.84VHH34 pKa = 7.26DD35 pKa = 3.98VTLKK39 pKa = 10.91FIEE42 pKa = 4.91DD43 pKa = 3.85CTTVYY48 pKa = 9.84TRR50 pKa = 11.84SLTKK54 pKa = 10.23CRR56 pKa = 11.84QEE58 pKa = 4.28RR59 pKa = 11.84LEE61 pKa = 4.67KK62 pKa = 10.27PAISPINQLKK72 pKa = 9.89RR73 pKa = 11.84LKK75 pKa = 9.34RR76 pKa = 11.84TYY78 pKa = 10.96GPIEE82 pKa = 4.06VKK84 pKa = 10.21EE85 pKa = 4.22DD86 pKa = 3.64CLISCEE92 pKa = 4.51DD93 pKa = 3.81EE94 pKa = 4.15SSHH97 pKa = 6.68RR98 pKa = 11.84QLDD101 pKa = 3.94DD102 pKa = 3.67SSLSCSEE109 pKa = 4.25TPFLNLMATKK119 pKa = 9.74EE120 pKa = 3.96LHH122 pKa = 4.86NTRR125 pKa = 11.84DD126 pKa = 3.48RR127 pKa = 11.84NQVHH131 pKa = 6.54GLHH134 pKa = 6.09SLNRR138 pKa = 11.84SFYY141 pKa = 8.8PLSYY145 pKa = 10.12KK146 pKa = 10.21IKK148 pKa = 10.65EE149 pKa = 3.79VDD151 pKa = 3.32QNLNTFDD158 pKa = 4.34NCWIDD163 pKa = 3.56QKK165 pKa = 11.35VKK167 pKa = 10.33EE168 pKa = 4.09SSYY171 pKa = 10.58YY172 pKa = 10.88DD173 pKa = 3.51KK174 pKa = 11.29LKK176 pKa = 10.99SDD178 pKa = 3.66TKK180 pKa = 10.14LTEE183 pKa = 3.88EE184 pKa = 5.15GEE186 pKa = 4.66FISQQDD192 pKa = 2.94ISMSKK197 pKa = 10.72YY198 pKa = 10.97SLDD201 pKa = 4.6KK202 pKa = 10.75INKK205 pKa = 8.76IYY207 pKa = 10.12PLSKK211 pKa = 9.92HH212 pKa = 5.91SSYY215 pKa = 11.58NNSVKK220 pKa = 10.43IGGAQSEE227 pKa = 4.45ISLNLQNSNLKK238 pKa = 10.46LGSNNGGLRR247 pKa = 11.84KK248 pKa = 9.02FAMLNNAMKK257 pKa = 10.34NQYY260 pKa = 9.8EE261 pKa = 4.09IAKK264 pKa = 9.53AKK266 pKa = 10.09CLNEE270 pKa = 3.95SKK272 pKa = 10.76DD273 pKa = 3.51IIKK276 pKa = 10.54EE277 pKa = 4.14SVRR280 pKa = 11.84VCSIVEE286 pKa = 3.84PLKK289 pKa = 10.93VRR291 pKa = 11.84TVTAEE296 pKa = 3.6EE297 pKa = 4.39ARR299 pKa = 11.84NQFIKK304 pKa = 10.59PIQKK308 pKa = 10.25LLWEE312 pKa = 5.12DD313 pKa = 3.88LLSHH317 pKa = 7.04EE318 pKa = 5.72DD319 pKa = 4.02FFLTRR324 pKa = 11.84NEE326 pKa = 4.26DD327 pKa = 5.06FEE329 pKa = 5.18SHH331 pKa = 5.4WNRR334 pKa = 11.84KK335 pKa = 7.92FKK337 pKa = 10.88FCDD340 pKa = 3.38LPYY343 pKa = 10.35HH344 pKa = 7.03ISGDD348 pKa = 3.81YY349 pKa = 10.19KK350 pKa = 11.14AATDD354 pKa = 3.66NLNRR358 pKa = 11.84NVINVVITQLSTLLPTNLIDD378 pKa = 6.1LFMKK382 pKa = 10.66NAGLHH387 pKa = 5.02YY388 pKa = 10.59LDD390 pKa = 4.29YY391 pKa = 11.62NEE393 pKa = 4.43VAFSLRR399 pKa = 11.84EE400 pKa = 4.07SPDD403 pKa = 3.47KK404 pKa = 11.18IGDD407 pKa = 3.52VYY409 pKa = 10.88LQKK412 pKa = 10.74NGQLMGSLTSFPLLCFINWISYY434 pKa = 8.03QFSKK438 pKa = 11.23YY439 pKa = 10.27LVNNYY444 pKa = 9.29PDD446 pKa = 3.34EE447 pKa = 4.59HH448 pKa = 8.31LSEE451 pKa = 4.48SCAINGDD458 pKa = 3.98DD459 pKa = 4.46IYY461 pKa = 11.4FKK463 pKa = 11.23ASLKK467 pKa = 10.71GYY469 pKa = 10.23AKK471 pKa = 8.86WQEE474 pKa = 4.17VVHH477 pKa = 6.4SFGLSPSPGKK487 pKa = 10.38NYY489 pKa = 10.39SSSNCFLINSRR500 pKa = 11.84NFLVNTKK507 pKa = 10.21SEE509 pKa = 4.18LKK511 pKa = 10.01EE512 pKa = 3.64IPFINFQMLPQYY524 pKa = 11.06SEE526 pKa = 4.15DD527 pKa = 3.67SNNKK531 pKa = 9.17FEE533 pKa = 4.84HH534 pKa = 5.93NKK536 pKa = 10.2KK537 pKa = 10.09EE538 pKa = 4.81LLNQMKK544 pKa = 10.17GDD546 pKa = 3.79SVYY549 pKa = 11.2SEE551 pKa = 4.42VNSPIGIIFRR561 pKa = 11.84TFCKK565 pKa = 9.67QANINLVKK573 pKa = 10.2PSKK576 pKa = 10.62FDD578 pKa = 3.1IQLIYY583 pKa = 10.86YY584 pKa = 8.43FYY586 pKa = 11.15SVWKK590 pKa = 10.37KK591 pKa = 10.66EE592 pKa = 3.98IIKK595 pKa = 10.28CPRR598 pKa = 11.84EE599 pKa = 3.94FYY601 pKa = 10.85LPSLIGGMGALTLDD615 pKa = 3.77EE616 pKa = 4.6FKK618 pKa = 10.27KK619 pKa = 9.61TNLLTKK625 pKa = 9.25TKK627 pKa = 10.23KK628 pKa = 10.48LSLLRR633 pKa = 11.84GFKK636 pKa = 10.39CKK638 pKa = 10.43SCFYY642 pKa = 8.95MQTYY646 pKa = 8.39EE647 pKa = 4.92QIGSDD652 pKa = 3.31KK653 pKa = 11.25DD654 pKa = 3.66QIEE657 pKa = 4.34SFAAKK662 pKa = 10.14LEE664 pKa = 4.15NIILDD669 pKa = 4.15EE670 pKa = 4.75LSGTDD675 pKa = 4.01PEE677 pKa = 4.33NKK679 pKa = 9.52QLSSKK684 pKa = 10.82SLLSPVSTFVSKK696 pKa = 11.12GGVEE700 pKa = 4.4KK701 pKa = 10.45YY702 pKa = 8.96TLQKK706 pKa = 10.53FFKK709 pKa = 10.18KK710 pKa = 10.16LGKK713 pKa = 10.12SSFRR717 pKa = 11.84NRR719 pKa = 11.84KK720 pKa = 8.4FKK722 pKa = 10.34PATVLSIEE730 pKa = 4.22KK731 pKa = 10.6SLL733 pKa = 3.9
Molecular weight: 84.36 kDa
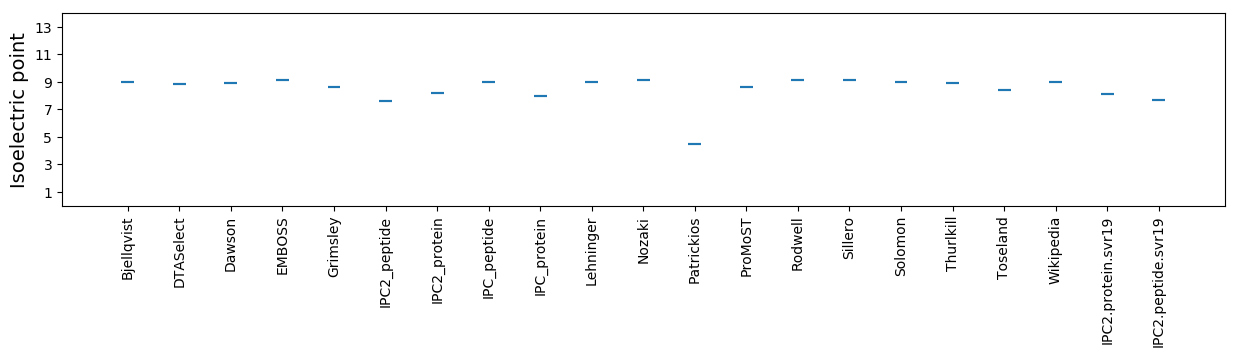
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KII2|A0A1L3KII2_9VIRU RNA-dependent RNA polymerase OS=Shahe narna-like virus 3 OX=1923431 PE=4 SV=1
MM1 pKa = 7.12FLKK4 pKa = 10.52RR5 pKa = 11.84SLAPVPRR12 pKa = 11.84SLIKK16 pKa = 10.4KK17 pKa = 9.94SRR19 pKa = 11.84DD20 pKa = 3.36DD21 pKa = 3.73HH22 pKa = 6.9KK23 pKa = 11.22STLTTPSEE31 pKa = 4.14RR32 pKa = 11.84VHH34 pKa = 7.26DD35 pKa = 3.98VTLKK39 pKa = 10.91FIEE42 pKa = 4.91DD43 pKa = 3.85CTTVYY48 pKa = 9.84TRR50 pKa = 11.84SLTKK54 pKa = 10.23CRR56 pKa = 11.84QEE58 pKa = 4.28RR59 pKa = 11.84LEE61 pKa = 4.67KK62 pKa = 10.27PAISPINQLKK72 pKa = 9.89RR73 pKa = 11.84LKK75 pKa = 9.34RR76 pKa = 11.84TYY78 pKa = 10.96GPIEE82 pKa = 4.06VKK84 pKa = 10.21EE85 pKa = 4.22DD86 pKa = 3.64CLISCEE92 pKa = 4.51DD93 pKa = 3.81EE94 pKa = 4.15SSHH97 pKa = 6.68RR98 pKa = 11.84QLDD101 pKa = 3.94DD102 pKa = 3.67SSLSCSEE109 pKa = 4.25TPFLNLMATKK119 pKa = 9.74EE120 pKa = 3.96LHH122 pKa = 4.86NTRR125 pKa = 11.84DD126 pKa = 3.48RR127 pKa = 11.84NQVHH131 pKa = 6.54GLHH134 pKa = 6.09SLNRR138 pKa = 11.84SFYY141 pKa = 8.8PLSYY145 pKa = 10.12KK146 pKa = 10.21IKK148 pKa = 10.65EE149 pKa = 3.79VDD151 pKa = 3.32QNLNTFDD158 pKa = 4.34NCWIDD163 pKa = 3.56QKK165 pKa = 11.35VKK167 pKa = 10.33EE168 pKa = 4.09SSYY171 pKa = 10.58YY172 pKa = 10.88DD173 pKa = 3.51KK174 pKa = 11.29LKK176 pKa = 10.99SDD178 pKa = 3.66TKK180 pKa = 10.14LTEE183 pKa = 3.88EE184 pKa = 5.15GEE186 pKa = 4.66FISQQDD192 pKa = 2.94ISMSKK197 pKa = 10.72YY198 pKa = 10.97SLDD201 pKa = 4.6KK202 pKa = 10.75INKK205 pKa = 8.76IYY207 pKa = 10.12PLSKK211 pKa = 9.92HH212 pKa = 5.91SSYY215 pKa = 11.58NNSVKK220 pKa = 10.43IGGAQSEE227 pKa = 4.45ISLNLQNSNLKK238 pKa = 10.46LGSNNGGLRR247 pKa = 11.84KK248 pKa = 9.02FAMLNNAMKK257 pKa = 10.34NQYY260 pKa = 9.8EE261 pKa = 4.09IAKK264 pKa = 9.53AKK266 pKa = 10.09CLNEE270 pKa = 3.95SKK272 pKa = 10.76DD273 pKa = 3.51IIKK276 pKa = 10.54EE277 pKa = 4.14SVRR280 pKa = 11.84VCSIVEE286 pKa = 3.84PLKK289 pKa = 10.93VRR291 pKa = 11.84TVTAEE296 pKa = 3.6EE297 pKa = 4.39ARR299 pKa = 11.84NQFIKK304 pKa = 10.59PIQKK308 pKa = 10.25LLWEE312 pKa = 5.12DD313 pKa = 3.88LLSHH317 pKa = 7.04EE318 pKa = 5.72DD319 pKa = 4.02FFLTRR324 pKa = 11.84NEE326 pKa = 4.26DD327 pKa = 5.06FEE329 pKa = 5.18SHH331 pKa = 5.4WNRR334 pKa = 11.84KK335 pKa = 7.92FKK337 pKa = 10.88FCDD340 pKa = 3.38LPYY343 pKa = 10.35HH344 pKa = 7.03ISGDD348 pKa = 3.81YY349 pKa = 10.19KK350 pKa = 11.14AATDD354 pKa = 3.66NLNRR358 pKa = 11.84NVINVVITQLSTLLPTNLIDD378 pKa = 6.1LFMKK382 pKa = 10.66NAGLHH387 pKa = 5.02YY388 pKa = 10.59LDD390 pKa = 4.29YY391 pKa = 11.62NEE393 pKa = 4.43VAFSLRR399 pKa = 11.84EE400 pKa = 4.07SPDD403 pKa = 3.47KK404 pKa = 11.18IGDD407 pKa = 3.52VYY409 pKa = 10.88LQKK412 pKa = 10.74NGQLMGSLTSFPLLCFINWISYY434 pKa = 8.03QFSKK438 pKa = 11.23YY439 pKa = 10.27LVNNYY444 pKa = 9.29PDD446 pKa = 3.34EE447 pKa = 4.59HH448 pKa = 8.31LSEE451 pKa = 4.48SCAINGDD458 pKa = 3.98DD459 pKa = 4.46IYY461 pKa = 11.4FKK463 pKa = 11.23ASLKK467 pKa = 10.71GYY469 pKa = 10.23AKK471 pKa = 8.86WQEE474 pKa = 4.17VVHH477 pKa = 6.4SFGLSPSPGKK487 pKa = 10.38NYY489 pKa = 10.39SSSNCFLINSRR500 pKa = 11.84NFLVNTKK507 pKa = 10.21SEE509 pKa = 4.18LKK511 pKa = 10.01EE512 pKa = 3.64IPFINFQMLPQYY524 pKa = 11.06SEE526 pKa = 4.15DD527 pKa = 3.67SNNKK531 pKa = 9.17FEE533 pKa = 4.84HH534 pKa = 5.93NKK536 pKa = 10.2KK537 pKa = 10.09EE538 pKa = 4.81LLNQMKK544 pKa = 10.17GDD546 pKa = 3.79SVYY549 pKa = 11.2SEE551 pKa = 4.42VNSPIGIIFRR561 pKa = 11.84TFCKK565 pKa = 9.67QANINLVKK573 pKa = 10.2PSKK576 pKa = 10.62FDD578 pKa = 3.1IQLIYY583 pKa = 10.86YY584 pKa = 8.43FYY586 pKa = 11.15SVWKK590 pKa = 10.37KK591 pKa = 10.66EE592 pKa = 3.98IIKK595 pKa = 10.28CPRR598 pKa = 11.84EE599 pKa = 3.94FYY601 pKa = 10.85LPSLIGGMGALTLDD615 pKa = 3.77EE616 pKa = 4.6FKK618 pKa = 10.27KK619 pKa = 9.61TNLLTKK625 pKa = 9.25TKK627 pKa = 10.23KK628 pKa = 10.48LSLLRR633 pKa = 11.84GFKK636 pKa = 10.39CKK638 pKa = 10.43SCFYY642 pKa = 8.95MQTYY646 pKa = 8.39EE647 pKa = 4.92QIGSDD652 pKa = 3.31KK653 pKa = 11.25DD654 pKa = 3.66QIEE657 pKa = 4.34SFAAKK662 pKa = 10.14LEE664 pKa = 4.15NIILDD669 pKa = 4.15EE670 pKa = 4.75LSGTDD675 pKa = 4.01PEE677 pKa = 4.33NKK679 pKa = 9.52QLSSKK684 pKa = 10.82SLLSPVSTFVSKK696 pKa = 11.12GGVEE700 pKa = 4.4KK701 pKa = 10.45YY702 pKa = 8.96TLQKK706 pKa = 10.53FFKK709 pKa = 10.18KK710 pKa = 10.16LGKK713 pKa = 10.12SSFRR717 pKa = 11.84NRR719 pKa = 11.84KK720 pKa = 8.4FKK722 pKa = 10.34PATVLSIEE730 pKa = 4.22KK731 pKa = 10.6SLL733 pKa = 3.9
MM1 pKa = 7.12FLKK4 pKa = 10.52RR5 pKa = 11.84SLAPVPRR12 pKa = 11.84SLIKK16 pKa = 10.4KK17 pKa = 9.94SRR19 pKa = 11.84DD20 pKa = 3.36DD21 pKa = 3.73HH22 pKa = 6.9KK23 pKa = 11.22STLTTPSEE31 pKa = 4.14RR32 pKa = 11.84VHH34 pKa = 7.26DD35 pKa = 3.98VTLKK39 pKa = 10.91FIEE42 pKa = 4.91DD43 pKa = 3.85CTTVYY48 pKa = 9.84TRR50 pKa = 11.84SLTKK54 pKa = 10.23CRR56 pKa = 11.84QEE58 pKa = 4.28RR59 pKa = 11.84LEE61 pKa = 4.67KK62 pKa = 10.27PAISPINQLKK72 pKa = 9.89RR73 pKa = 11.84LKK75 pKa = 9.34RR76 pKa = 11.84TYY78 pKa = 10.96GPIEE82 pKa = 4.06VKK84 pKa = 10.21EE85 pKa = 4.22DD86 pKa = 3.64CLISCEE92 pKa = 4.51DD93 pKa = 3.81EE94 pKa = 4.15SSHH97 pKa = 6.68RR98 pKa = 11.84QLDD101 pKa = 3.94DD102 pKa = 3.67SSLSCSEE109 pKa = 4.25TPFLNLMATKK119 pKa = 9.74EE120 pKa = 3.96LHH122 pKa = 4.86NTRR125 pKa = 11.84DD126 pKa = 3.48RR127 pKa = 11.84NQVHH131 pKa = 6.54GLHH134 pKa = 6.09SLNRR138 pKa = 11.84SFYY141 pKa = 8.8PLSYY145 pKa = 10.12KK146 pKa = 10.21IKK148 pKa = 10.65EE149 pKa = 3.79VDD151 pKa = 3.32QNLNTFDD158 pKa = 4.34NCWIDD163 pKa = 3.56QKK165 pKa = 11.35VKK167 pKa = 10.33EE168 pKa = 4.09SSYY171 pKa = 10.58YY172 pKa = 10.88DD173 pKa = 3.51KK174 pKa = 11.29LKK176 pKa = 10.99SDD178 pKa = 3.66TKK180 pKa = 10.14LTEE183 pKa = 3.88EE184 pKa = 5.15GEE186 pKa = 4.66FISQQDD192 pKa = 2.94ISMSKK197 pKa = 10.72YY198 pKa = 10.97SLDD201 pKa = 4.6KK202 pKa = 10.75INKK205 pKa = 8.76IYY207 pKa = 10.12PLSKK211 pKa = 9.92HH212 pKa = 5.91SSYY215 pKa = 11.58NNSVKK220 pKa = 10.43IGGAQSEE227 pKa = 4.45ISLNLQNSNLKK238 pKa = 10.46LGSNNGGLRR247 pKa = 11.84KK248 pKa = 9.02FAMLNNAMKK257 pKa = 10.34NQYY260 pKa = 9.8EE261 pKa = 4.09IAKK264 pKa = 9.53AKK266 pKa = 10.09CLNEE270 pKa = 3.95SKK272 pKa = 10.76DD273 pKa = 3.51IIKK276 pKa = 10.54EE277 pKa = 4.14SVRR280 pKa = 11.84VCSIVEE286 pKa = 3.84PLKK289 pKa = 10.93VRR291 pKa = 11.84TVTAEE296 pKa = 3.6EE297 pKa = 4.39ARR299 pKa = 11.84NQFIKK304 pKa = 10.59PIQKK308 pKa = 10.25LLWEE312 pKa = 5.12DD313 pKa = 3.88LLSHH317 pKa = 7.04EE318 pKa = 5.72DD319 pKa = 4.02FFLTRR324 pKa = 11.84NEE326 pKa = 4.26DD327 pKa = 5.06FEE329 pKa = 5.18SHH331 pKa = 5.4WNRR334 pKa = 11.84KK335 pKa = 7.92FKK337 pKa = 10.88FCDD340 pKa = 3.38LPYY343 pKa = 10.35HH344 pKa = 7.03ISGDD348 pKa = 3.81YY349 pKa = 10.19KK350 pKa = 11.14AATDD354 pKa = 3.66NLNRR358 pKa = 11.84NVINVVITQLSTLLPTNLIDD378 pKa = 6.1LFMKK382 pKa = 10.66NAGLHH387 pKa = 5.02YY388 pKa = 10.59LDD390 pKa = 4.29YY391 pKa = 11.62NEE393 pKa = 4.43VAFSLRR399 pKa = 11.84EE400 pKa = 4.07SPDD403 pKa = 3.47KK404 pKa = 11.18IGDD407 pKa = 3.52VYY409 pKa = 10.88LQKK412 pKa = 10.74NGQLMGSLTSFPLLCFINWISYY434 pKa = 8.03QFSKK438 pKa = 11.23YY439 pKa = 10.27LVNNYY444 pKa = 9.29PDD446 pKa = 3.34EE447 pKa = 4.59HH448 pKa = 8.31LSEE451 pKa = 4.48SCAINGDD458 pKa = 3.98DD459 pKa = 4.46IYY461 pKa = 11.4FKK463 pKa = 11.23ASLKK467 pKa = 10.71GYY469 pKa = 10.23AKK471 pKa = 8.86WQEE474 pKa = 4.17VVHH477 pKa = 6.4SFGLSPSPGKK487 pKa = 10.38NYY489 pKa = 10.39SSSNCFLINSRR500 pKa = 11.84NFLVNTKK507 pKa = 10.21SEE509 pKa = 4.18LKK511 pKa = 10.01EE512 pKa = 3.64IPFINFQMLPQYY524 pKa = 11.06SEE526 pKa = 4.15DD527 pKa = 3.67SNNKK531 pKa = 9.17FEE533 pKa = 4.84HH534 pKa = 5.93NKK536 pKa = 10.2KK537 pKa = 10.09EE538 pKa = 4.81LLNQMKK544 pKa = 10.17GDD546 pKa = 3.79SVYY549 pKa = 11.2SEE551 pKa = 4.42VNSPIGIIFRR561 pKa = 11.84TFCKK565 pKa = 9.67QANINLVKK573 pKa = 10.2PSKK576 pKa = 10.62FDD578 pKa = 3.1IQLIYY583 pKa = 10.86YY584 pKa = 8.43FYY586 pKa = 11.15SVWKK590 pKa = 10.37KK591 pKa = 10.66EE592 pKa = 3.98IIKK595 pKa = 10.28CPRR598 pKa = 11.84EE599 pKa = 3.94FYY601 pKa = 10.85LPSLIGGMGALTLDD615 pKa = 3.77EE616 pKa = 4.6FKK618 pKa = 10.27KK619 pKa = 9.61TNLLTKK625 pKa = 9.25TKK627 pKa = 10.23KK628 pKa = 10.48LSLLRR633 pKa = 11.84GFKK636 pKa = 10.39CKK638 pKa = 10.43SCFYY642 pKa = 8.95MQTYY646 pKa = 8.39EE647 pKa = 4.92QIGSDD652 pKa = 3.31KK653 pKa = 11.25DD654 pKa = 3.66QIEE657 pKa = 4.34SFAAKK662 pKa = 10.14LEE664 pKa = 4.15NIILDD669 pKa = 4.15EE670 pKa = 4.75LSGTDD675 pKa = 4.01PEE677 pKa = 4.33NKK679 pKa = 9.52QLSSKK684 pKa = 10.82SLLSPVSTFVSKK696 pKa = 11.12GGVEE700 pKa = 4.4KK701 pKa = 10.45YY702 pKa = 8.96TLQKK706 pKa = 10.53FFKK709 pKa = 10.18KK710 pKa = 10.16LGKK713 pKa = 10.12SSFRR717 pKa = 11.84NRR719 pKa = 11.84KK720 pKa = 8.4FKK722 pKa = 10.34PATVLSIEE730 pKa = 4.22KK731 pKa = 10.6SLL733 pKa = 3.9
Molecular weight: 84.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
733 |
733 |
733 |
733.0 |
84.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.001 ± 0.0 | 2.183 ± 0.0 |
5.184 ± 0.0 | 6.412 ± 0.0 |
5.321 ± 0.0 | 3.82 ± 0.0 |
1.91 ± 0.0 | 6.276 ± 0.0 |
10.368 ± 0.0 | 11.187 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.501 ± 0.0 | 7.094 ± 0.0 |
3.683 ± 0.0 | 3.82 ± 0.0 |
3.683 ± 0.0 | 10.778 ± 0.0 |
4.638 ± 0.0 | 4.229 ± 0.0 |
0.819 ± 0.0 | 4.093 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
