
Methylovorus sp. MM2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Methylophilaceae; Methylovorus; unclassified Methylovorus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
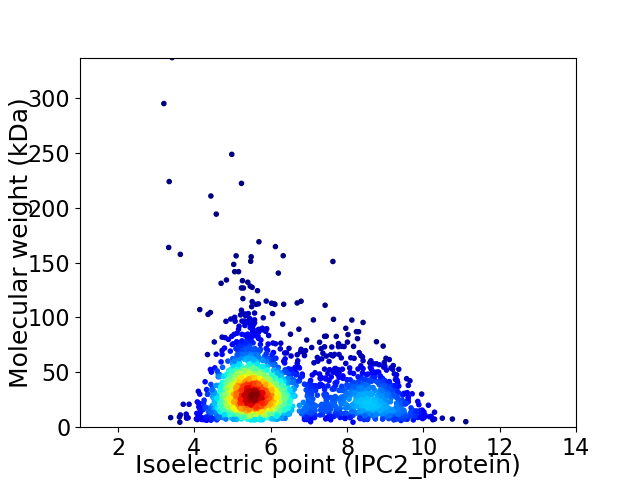
Virtual 2D-PAGE plot for 2256 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A9T466|A0A1A9T466_9PROT Uncharacterized protein OS=Methylovorus sp. MM2 OX=1848038 GN=A7981_04980 PE=4 SV=1
MM1 pKa = 7.45PTFISNFLSSDD12 pKa = 4.27AIRR15 pKa = 11.84EE16 pKa = 4.18AFGGIDD22 pKa = 3.4AVDD25 pKa = 3.61EE26 pKa = 4.19QMEE29 pKa = 4.61SLGEE33 pKa = 4.04ALDD36 pKa = 3.64IAFSDD41 pKa = 4.56EE42 pKa = 4.33YY43 pKa = 10.98LGPAYY48 pKa = 9.62EE49 pKa = 4.43VVPSSILASNNAMSGRR65 pKa = 11.84LVNGDD70 pKa = 3.68SFNITGKK77 pKa = 10.2EE78 pKa = 3.71LLADD82 pKa = 3.71YY83 pKa = 9.54PHH85 pKa = 7.47IYY87 pKa = 10.11SFIYY91 pKa = 10.13RR92 pKa = 11.84FHH94 pKa = 7.07QGGSVSFSGDD104 pKa = 2.61MSYY107 pKa = 9.21YY108 pKa = 10.15TNPNNLPGYY117 pKa = 9.64EE118 pKa = 4.07NTFDD122 pKa = 4.16GYY124 pKa = 10.21VSKK127 pKa = 11.16LIIEE131 pKa = 4.63SGQGSKK137 pKa = 7.49VTYY140 pKa = 9.66IGRR143 pKa = 11.84GNIEE147 pKa = 4.32SNGDD151 pKa = 3.11INEE154 pKa = 4.24FVATSMQLQIGKK166 pKa = 10.19LSLVVNGEE174 pKa = 4.09INLSFVDD181 pKa = 5.76DD182 pKa = 4.1IDD184 pKa = 4.21DD185 pKa = 3.38VSGIVTGFNLTSGKK199 pKa = 10.57YY200 pKa = 8.37SFSMSGLVLDD210 pKa = 5.37AVDD213 pKa = 4.54FDD215 pKa = 5.06AYY217 pKa = 10.91SEE219 pKa = 4.42SGFDD223 pKa = 4.78FLDD226 pKa = 3.65NLLAGNDD233 pKa = 3.81TLDD236 pKa = 3.96GNVSNNMIYY245 pKa = 10.91GLVGDD250 pKa = 4.95DD251 pKa = 5.06SINGLSGNDD260 pKa = 3.01ILYY263 pKa = 10.66GGEE266 pKa = 4.17GSDD269 pKa = 3.62TLVGGIGNDD278 pKa = 3.56TLDD281 pKa = 4.49GGDD284 pKa = 4.23GDD286 pKa = 4.59DD287 pKa = 4.45SLVGGAGNDD296 pKa = 3.5VYY298 pKa = 11.0IVGEE302 pKa = 3.94FGDD305 pKa = 4.44IVDD308 pKa = 4.0EE309 pKa = 4.77TISGSSGLDD318 pKa = 2.98RR319 pKa = 11.84VKK321 pKa = 10.89LGSTYY326 pKa = 11.62DD327 pKa = 3.33MTHH330 pKa = 6.62YY331 pKa = 11.2DD332 pKa = 3.19LTDD335 pKa = 3.26NVEE338 pKa = 4.25NLDD341 pKa = 3.63ASAVTTVWPSTWLEE355 pKa = 3.97LYY357 pKa = 10.69GNSLANVIIGSSSNDD372 pKa = 3.22YY373 pKa = 11.24LIGLDD378 pKa = 4.91GNDD381 pKa = 3.77TLIGGAGDD389 pKa = 5.17DD390 pKa = 4.06ILLGGAGVDD399 pKa = 3.7NLQGGAGNDD408 pKa = 3.65TYY410 pKa = 11.75NVNLKK415 pKa = 9.64TIGTGVNITAALEE428 pKa = 3.97DD429 pKa = 4.94TITDD433 pKa = 3.7TAGTADD439 pKa = 3.85TLVLLGGLGANKK451 pKa = 9.63TSTITLVAALEE462 pKa = 4.13NLDD465 pKa = 3.85ASSAEE470 pKa = 4.06VNNTIVGAVALNLTGNASNNEE491 pKa = 4.17IIGSNGKK498 pKa = 7.99NTILGLAGNDD508 pKa = 4.08TIDD511 pKa = 4.62GLDD514 pKa = 3.95GDD516 pKa = 4.93DD517 pKa = 5.18SIDD520 pKa = 3.55GGVGNDD526 pKa = 3.75SLSGGDD532 pKa = 3.8GDD534 pKa = 4.04DD535 pKa = 3.87TYY537 pKa = 12.11VVALKK542 pKa = 10.56LQGSGAGANVIIEE555 pKa = 4.17DD556 pKa = 4.61TIDD559 pKa = 3.25EE560 pKa = 4.55ALDD563 pKa = 3.46EE564 pKa = 4.74GRR566 pKa = 11.84DD567 pKa = 3.75TLRR570 pKa = 11.84LAGSAKK576 pKa = 9.05LTNASTIEE584 pKa = 4.03LTGDD588 pKa = 3.88LANIEE593 pKa = 4.79NIDD596 pKa = 3.58ASLTGSTLLNLIGNGSSNYY615 pKa = 10.43LIGNSAANWLDD626 pKa = 3.6GGDD629 pKa = 4.59GDD631 pKa = 5.08DD632 pKa = 4.22TLDD635 pKa = 3.89GGGGANTMIGGFGNDD650 pKa = 2.8TYY652 pKa = 11.73VVNNAKK658 pKa = 10.45DD659 pKa = 3.66VVFDD663 pKa = 3.84GVNPSILRR671 pKa = 11.84LSIDD675 pKa = 3.54SQGNEE680 pKa = 3.71ANTGGFNAIYY690 pKa = 9.86SINDD694 pKa = 3.27RR695 pKa = 11.84YY696 pKa = 11.18VIFVSGASNLVSNDD710 pKa = 3.2NNLSADD716 pKa = 3.7VFIKK720 pKa = 10.47DD721 pKa = 3.33IQTGNVTRR729 pKa = 11.84VSTNSADD736 pKa = 3.46QEE738 pKa = 4.77SNHH741 pKa = 5.91VYY743 pKa = 10.67DD744 pKa = 5.27AVPEE748 pKa = 4.07LAISLDD754 pKa = 3.1GKK756 pKa = 10.43YY757 pKa = 10.57LVFSTTDD764 pKa = 2.97TGLVNGDD771 pKa = 3.53VNDD774 pKa = 3.77TRR776 pKa = 11.84DD777 pKa = 3.24IYY779 pKa = 11.35LKK781 pKa = 10.52NLQTGEE787 pKa = 4.01IKK789 pKa = 10.84LVDD792 pKa = 3.58VSINGAHH799 pKa = 6.43TDD801 pKa = 3.13SHH803 pKa = 7.85GYY805 pKa = 10.48EE806 pKa = 4.08SFNPSFSPDD815 pKa = 3.19SKK817 pKa = 11.34SVLFSSFDD825 pKa = 3.61PLLVNNGNASNVGVYY840 pKa = 9.82IKK842 pKa = 10.52NIQTGAVKK850 pKa = 10.36LVSTNSLGDD859 pKa = 3.62SANDD863 pKa = 3.04ATYY866 pKa = 10.36AYY868 pKa = 9.99GYY870 pKa = 10.67SSDD873 pKa = 3.39GKK875 pKa = 10.75KK876 pKa = 9.8ILLVSSANNFTVGDD890 pKa = 3.97SNGFQDD896 pKa = 4.97VFIKK900 pKa = 10.64NLQTNAVQLINTDD913 pKa = 3.32KK914 pKa = 11.39NGNQANNHH922 pKa = 5.84SYY924 pKa = 11.4NPLFSKK930 pKa = 10.63DD931 pKa = 2.89GNYY934 pKa = 9.69VVFEE938 pKa = 4.26SLASNLVEE946 pKa = 4.8EE947 pKa = 4.44YY948 pKa = 10.39TDD950 pKa = 3.58YY951 pKa = 11.67GKK953 pKa = 10.67LQIYY957 pKa = 9.62IKK959 pKa = 10.46NLQTGDD965 pKa = 3.21IQLVSKK971 pKa = 11.33NEE973 pKa = 4.05LGEE976 pKa = 4.2AGYY979 pKa = 11.15GDD981 pKa = 4.52SLNASISSNGRR992 pKa = 11.84YY993 pKa = 7.99VVFMSSASNLVVGDD1007 pKa = 3.91TNGMLDD1013 pKa = 3.38VFVRR1017 pKa = 11.84DD1018 pKa = 4.12LYY1020 pKa = 9.43TNEE1023 pKa = 3.83VKK1025 pKa = 10.59RR1026 pKa = 11.84ISVSLNGQEE1035 pKa = 4.64GNGDD1039 pKa = 3.86SYY1041 pKa = 11.83TNSALLGGSTFSSDD1055 pKa = 2.42GGYY1058 pKa = 10.12ILFHH1062 pKa = 6.91SNASNLDD1069 pKa = 3.3ASDD1072 pKa = 3.95NNSVADD1078 pKa = 3.72VFRR1081 pKa = 11.84VANPFLIADD1090 pKa = 5.45GIDD1093 pKa = 3.14IVIAAISYY1101 pKa = 8.21TLGADD1106 pKa = 3.43IEE1108 pKa = 4.3NLTLSGTSGLSGRR1121 pKa = 11.84GNDD1124 pKa = 3.96LDD1126 pKa = 4.54NVITGNSGANKK1137 pKa = 9.68LQSLVGNDD1145 pKa = 3.6TLLGGAGSDD1154 pKa = 3.72TLDD1157 pKa = 3.68GGLGIDD1163 pKa = 4.22SMEE1166 pKa = 4.08GSAGNDD1172 pKa = 3.21VYY1174 pKa = 11.02IVDD1177 pKa = 5.6DD1178 pKa = 4.04INDD1181 pKa = 4.09LVVEE1185 pKa = 4.23TDD1187 pKa = 3.57VNSKK1191 pKa = 10.6SGGIDD1196 pKa = 2.96TVRR1199 pKa = 11.84IANTFIGAYY1208 pKa = 8.5YY1209 pKa = 10.9ALIANVEE1216 pKa = 4.12NLDD1219 pKa = 3.33GSAYY1223 pKa = 9.77ISGIDD1228 pKa = 3.44LRR1230 pKa = 11.84GNGLVNTITGGIGNDD1245 pKa = 3.51TLAGSAYY1252 pKa = 8.75EE1253 pKa = 3.87TDD1255 pKa = 3.37QYY1257 pKa = 11.66INDD1260 pKa = 3.99GLVDD1264 pKa = 3.61TFKK1267 pKa = 11.04GGKK1270 pKa = 9.95GDD1272 pKa = 3.64DD1273 pKa = 3.87TYY1275 pKa = 11.69HH1276 pKa = 6.76VFLQTTGSANKK1287 pKa = 8.89ATVSIQDD1294 pKa = 4.26SIVEE1298 pKa = 4.29SANQGTDD1305 pKa = 2.95TVILHH1310 pKa = 6.7GSDD1313 pKa = 3.45NNTLTKK1319 pKa = 10.38ASKK1322 pKa = 8.69ITLTANLEE1330 pKa = 4.15HH1331 pKa = 7.3LDD1333 pKa = 4.52ASDD1336 pKa = 5.08TDD1338 pKa = 4.42STWLNLTGNVASNQITGNDD1357 pKa = 3.4GHH1359 pKa = 7.29NIILGLGGNDD1369 pKa = 2.87ILYY1372 pKa = 10.47GGNGHH1377 pKa = 6.84DD1378 pKa = 4.31TLDD1381 pKa = 3.69GGIGNDD1387 pKa = 3.72VLNGGADD1394 pKa = 3.75DD1395 pKa = 4.95DD1396 pKa = 4.49KK1397 pKa = 11.78LIGGLGNDD1405 pKa = 3.97TLTGGNGSDD1414 pKa = 2.5IFLFNTALNATTNVDD1429 pKa = 3.82TITDD1433 pKa = 4.13FTSGEE1438 pKa = 3.86DD1439 pKa = 4.23HH1440 pKa = 7.3LYY1442 pKa = 10.7LDD1444 pKa = 3.51NAIFKK1449 pKa = 10.78KK1450 pKa = 9.5LTNEE1454 pKa = 4.19SALSADD1460 pKa = 3.08NFYY1463 pKa = 11.48VVGSGAQTAHH1473 pKa = 5.85QFIIYY1478 pKa = 10.22DD1479 pKa = 3.79GASGNLFYY1487 pKa = 11.13DD1488 pKa = 3.72ADD1490 pKa = 4.25GSGLLAPVLFANLEE1504 pKa = 4.14SAPALTAGDD1513 pKa = 3.72IQIVV1517 pKa = 3.37
MM1 pKa = 7.45PTFISNFLSSDD12 pKa = 4.27AIRR15 pKa = 11.84EE16 pKa = 4.18AFGGIDD22 pKa = 3.4AVDD25 pKa = 3.61EE26 pKa = 4.19QMEE29 pKa = 4.61SLGEE33 pKa = 4.04ALDD36 pKa = 3.64IAFSDD41 pKa = 4.56EE42 pKa = 4.33YY43 pKa = 10.98LGPAYY48 pKa = 9.62EE49 pKa = 4.43VVPSSILASNNAMSGRR65 pKa = 11.84LVNGDD70 pKa = 3.68SFNITGKK77 pKa = 10.2EE78 pKa = 3.71LLADD82 pKa = 3.71YY83 pKa = 9.54PHH85 pKa = 7.47IYY87 pKa = 10.11SFIYY91 pKa = 10.13RR92 pKa = 11.84FHH94 pKa = 7.07QGGSVSFSGDD104 pKa = 2.61MSYY107 pKa = 9.21YY108 pKa = 10.15TNPNNLPGYY117 pKa = 9.64EE118 pKa = 4.07NTFDD122 pKa = 4.16GYY124 pKa = 10.21VSKK127 pKa = 11.16LIIEE131 pKa = 4.63SGQGSKK137 pKa = 7.49VTYY140 pKa = 9.66IGRR143 pKa = 11.84GNIEE147 pKa = 4.32SNGDD151 pKa = 3.11INEE154 pKa = 4.24FVATSMQLQIGKK166 pKa = 10.19LSLVVNGEE174 pKa = 4.09INLSFVDD181 pKa = 5.76DD182 pKa = 4.1IDD184 pKa = 4.21DD185 pKa = 3.38VSGIVTGFNLTSGKK199 pKa = 10.57YY200 pKa = 8.37SFSMSGLVLDD210 pKa = 5.37AVDD213 pKa = 4.54FDD215 pKa = 5.06AYY217 pKa = 10.91SEE219 pKa = 4.42SGFDD223 pKa = 4.78FLDD226 pKa = 3.65NLLAGNDD233 pKa = 3.81TLDD236 pKa = 3.96GNVSNNMIYY245 pKa = 10.91GLVGDD250 pKa = 4.95DD251 pKa = 5.06SINGLSGNDD260 pKa = 3.01ILYY263 pKa = 10.66GGEE266 pKa = 4.17GSDD269 pKa = 3.62TLVGGIGNDD278 pKa = 3.56TLDD281 pKa = 4.49GGDD284 pKa = 4.23GDD286 pKa = 4.59DD287 pKa = 4.45SLVGGAGNDD296 pKa = 3.5VYY298 pKa = 11.0IVGEE302 pKa = 3.94FGDD305 pKa = 4.44IVDD308 pKa = 4.0EE309 pKa = 4.77TISGSSGLDD318 pKa = 2.98RR319 pKa = 11.84VKK321 pKa = 10.89LGSTYY326 pKa = 11.62DD327 pKa = 3.33MTHH330 pKa = 6.62YY331 pKa = 11.2DD332 pKa = 3.19LTDD335 pKa = 3.26NVEE338 pKa = 4.25NLDD341 pKa = 3.63ASAVTTVWPSTWLEE355 pKa = 3.97LYY357 pKa = 10.69GNSLANVIIGSSSNDD372 pKa = 3.22YY373 pKa = 11.24LIGLDD378 pKa = 4.91GNDD381 pKa = 3.77TLIGGAGDD389 pKa = 5.17DD390 pKa = 4.06ILLGGAGVDD399 pKa = 3.7NLQGGAGNDD408 pKa = 3.65TYY410 pKa = 11.75NVNLKK415 pKa = 9.64TIGTGVNITAALEE428 pKa = 3.97DD429 pKa = 4.94TITDD433 pKa = 3.7TAGTADD439 pKa = 3.85TLVLLGGLGANKK451 pKa = 9.63TSTITLVAALEE462 pKa = 4.13NLDD465 pKa = 3.85ASSAEE470 pKa = 4.06VNNTIVGAVALNLTGNASNNEE491 pKa = 4.17IIGSNGKK498 pKa = 7.99NTILGLAGNDD508 pKa = 4.08TIDD511 pKa = 4.62GLDD514 pKa = 3.95GDD516 pKa = 4.93DD517 pKa = 5.18SIDD520 pKa = 3.55GGVGNDD526 pKa = 3.75SLSGGDD532 pKa = 3.8GDD534 pKa = 4.04DD535 pKa = 3.87TYY537 pKa = 12.11VVALKK542 pKa = 10.56LQGSGAGANVIIEE555 pKa = 4.17DD556 pKa = 4.61TIDD559 pKa = 3.25EE560 pKa = 4.55ALDD563 pKa = 3.46EE564 pKa = 4.74GRR566 pKa = 11.84DD567 pKa = 3.75TLRR570 pKa = 11.84LAGSAKK576 pKa = 9.05LTNASTIEE584 pKa = 4.03LTGDD588 pKa = 3.88LANIEE593 pKa = 4.79NIDD596 pKa = 3.58ASLTGSTLLNLIGNGSSNYY615 pKa = 10.43LIGNSAANWLDD626 pKa = 3.6GGDD629 pKa = 4.59GDD631 pKa = 5.08DD632 pKa = 4.22TLDD635 pKa = 3.89GGGGANTMIGGFGNDD650 pKa = 2.8TYY652 pKa = 11.73VVNNAKK658 pKa = 10.45DD659 pKa = 3.66VVFDD663 pKa = 3.84GVNPSILRR671 pKa = 11.84LSIDD675 pKa = 3.54SQGNEE680 pKa = 3.71ANTGGFNAIYY690 pKa = 9.86SINDD694 pKa = 3.27RR695 pKa = 11.84YY696 pKa = 11.18VIFVSGASNLVSNDD710 pKa = 3.2NNLSADD716 pKa = 3.7VFIKK720 pKa = 10.47DD721 pKa = 3.33IQTGNVTRR729 pKa = 11.84VSTNSADD736 pKa = 3.46QEE738 pKa = 4.77SNHH741 pKa = 5.91VYY743 pKa = 10.67DD744 pKa = 5.27AVPEE748 pKa = 4.07LAISLDD754 pKa = 3.1GKK756 pKa = 10.43YY757 pKa = 10.57LVFSTTDD764 pKa = 2.97TGLVNGDD771 pKa = 3.53VNDD774 pKa = 3.77TRR776 pKa = 11.84DD777 pKa = 3.24IYY779 pKa = 11.35LKK781 pKa = 10.52NLQTGEE787 pKa = 4.01IKK789 pKa = 10.84LVDD792 pKa = 3.58VSINGAHH799 pKa = 6.43TDD801 pKa = 3.13SHH803 pKa = 7.85GYY805 pKa = 10.48EE806 pKa = 4.08SFNPSFSPDD815 pKa = 3.19SKK817 pKa = 11.34SVLFSSFDD825 pKa = 3.61PLLVNNGNASNVGVYY840 pKa = 9.82IKK842 pKa = 10.52NIQTGAVKK850 pKa = 10.36LVSTNSLGDD859 pKa = 3.62SANDD863 pKa = 3.04ATYY866 pKa = 10.36AYY868 pKa = 9.99GYY870 pKa = 10.67SSDD873 pKa = 3.39GKK875 pKa = 10.75KK876 pKa = 9.8ILLVSSANNFTVGDD890 pKa = 3.97SNGFQDD896 pKa = 4.97VFIKK900 pKa = 10.64NLQTNAVQLINTDD913 pKa = 3.32KK914 pKa = 11.39NGNQANNHH922 pKa = 5.84SYY924 pKa = 11.4NPLFSKK930 pKa = 10.63DD931 pKa = 2.89GNYY934 pKa = 9.69VVFEE938 pKa = 4.26SLASNLVEE946 pKa = 4.8EE947 pKa = 4.44YY948 pKa = 10.39TDD950 pKa = 3.58YY951 pKa = 11.67GKK953 pKa = 10.67LQIYY957 pKa = 9.62IKK959 pKa = 10.46NLQTGDD965 pKa = 3.21IQLVSKK971 pKa = 11.33NEE973 pKa = 4.05LGEE976 pKa = 4.2AGYY979 pKa = 11.15GDD981 pKa = 4.52SLNASISSNGRR992 pKa = 11.84YY993 pKa = 7.99VVFMSSASNLVVGDD1007 pKa = 3.91TNGMLDD1013 pKa = 3.38VFVRR1017 pKa = 11.84DD1018 pKa = 4.12LYY1020 pKa = 9.43TNEE1023 pKa = 3.83VKK1025 pKa = 10.59RR1026 pKa = 11.84ISVSLNGQEE1035 pKa = 4.64GNGDD1039 pKa = 3.86SYY1041 pKa = 11.83TNSALLGGSTFSSDD1055 pKa = 2.42GGYY1058 pKa = 10.12ILFHH1062 pKa = 6.91SNASNLDD1069 pKa = 3.3ASDD1072 pKa = 3.95NNSVADD1078 pKa = 3.72VFRR1081 pKa = 11.84VANPFLIADD1090 pKa = 5.45GIDD1093 pKa = 3.14IVIAAISYY1101 pKa = 8.21TLGADD1106 pKa = 3.43IEE1108 pKa = 4.3NLTLSGTSGLSGRR1121 pKa = 11.84GNDD1124 pKa = 3.96LDD1126 pKa = 4.54NVITGNSGANKK1137 pKa = 9.68LQSLVGNDD1145 pKa = 3.6TLLGGAGSDD1154 pKa = 3.72TLDD1157 pKa = 3.68GGLGIDD1163 pKa = 4.22SMEE1166 pKa = 4.08GSAGNDD1172 pKa = 3.21VYY1174 pKa = 11.02IVDD1177 pKa = 5.6DD1178 pKa = 4.04INDD1181 pKa = 4.09LVVEE1185 pKa = 4.23TDD1187 pKa = 3.57VNSKK1191 pKa = 10.6SGGIDD1196 pKa = 2.96TVRR1199 pKa = 11.84IANTFIGAYY1208 pKa = 8.5YY1209 pKa = 10.9ALIANVEE1216 pKa = 4.12NLDD1219 pKa = 3.33GSAYY1223 pKa = 9.77ISGIDD1228 pKa = 3.44LRR1230 pKa = 11.84GNGLVNTITGGIGNDD1245 pKa = 3.51TLAGSAYY1252 pKa = 8.75EE1253 pKa = 3.87TDD1255 pKa = 3.37QYY1257 pKa = 11.66INDD1260 pKa = 3.99GLVDD1264 pKa = 3.61TFKK1267 pKa = 11.04GGKK1270 pKa = 9.95GDD1272 pKa = 3.64DD1273 pKa = 3.87TYY1275 pKa = 11.69HH1276 pKa = 6.76VFLQTTGSANKK1287 pKa = 8.89ATVSIQDD1294 pKa = 4.26SIVEE1298 pKa = 4.29SANQGTDD1305 pKa = 2.95TVILHH1310 pKa = 6.7GSDD1313 pKa = 3.45NNTLTKK1319 pKa = 10.38ASKK1322 pKa = 8.69ITLTANLEE1330 pKa = 4.15HH1331 pKa = 7.3LDD1333 pKa = 4.52ASDD1336 pKa = 5.08TDD1338 pKa = 4.42STWLNLTGNVASNQITGNDD1357 pKa = 3.4GHH1359 pKa = 7.29NIILGLGGNDD1369 pKa = 2.87ILYY1372 pKa = 10.47GGNGHH1377 pKa = 6.84DD1378 pKa = 4.31TLDD1381 pKa = 3.69GGIGNDD1387 pKa = 3.72VLNGGADD1394 pKa = 3.75DD1395 pKa = 4.95DD1396 pKa = 4.49KK1397 pKa = 11.78LIGGLGNDD1405 pKa = 3.97TLTGGNGSDD1414 pKa = 2.5IFLFNTALNATTNVDD1429 pKa = 3.82TITDD1433 pKa = 4.13FTSGEE1438 pKa = 3.86DD1439 pKa = 4.23HH1440 pKa = 7.3LYY1442 pKa = 10.7LDD1444 pKa = 3.51NAIFKK1449 pKa = 10.78KK1450 pKa = 9.5LTNEE1454 pKa = 4.19SALSADD1460 pKa = 3.08NFYY1463 pKa = 11.48VVGSGAQTAHH1473 pKa = 5.85QFIIYY1478 pKa = 10.22DD1479 pKa = 3.79GASGNLFYY1487 pKa = 11.13DD1488 pKa = 3.72ADD1490 pKa = 4.25GSGLLAPVLFANLEE1504 pKa = 4.14SAPALTAGDD1513 pKa = 3.72IQIVV1517 pKa = 3.37
Molecular weight: 157.48 kDa
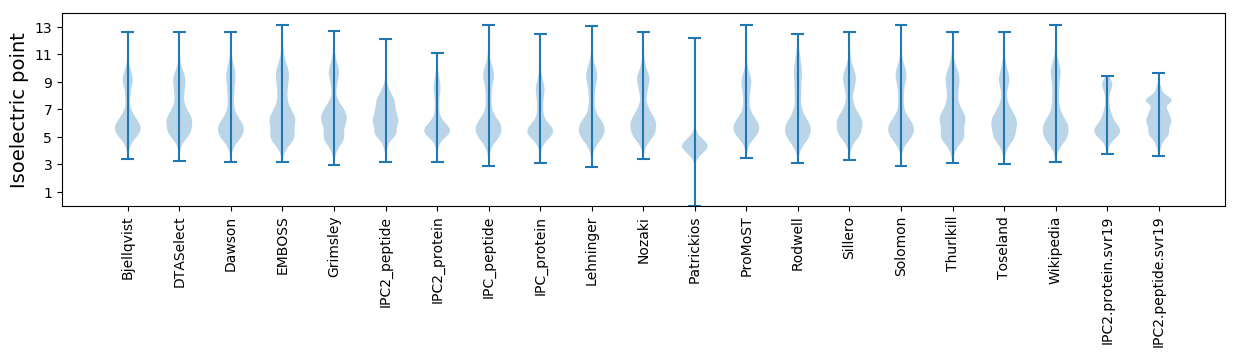
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A9T200|A0A1A9T200_9PROT Uncharacterized protein OS=Methylovorus sp. MM2 OX=1848038 GN=A7981_00760 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 8.87VRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.65GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.17GRR39 pKa = 11.84ARR41 pKa = 11.84LAVV44 pKa = 3.42
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 8.87VRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.65GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.17GRR39 pKa = 11.84ARR41 pKa = 11.84LAVV44 pKa = 3.42
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
731518 |
39 |
3267 |
324.3 |
35.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.725 ± 0.058 | 0.78 ± 0.018 |
5.6 ± 0.048 | 5.793 ± 0.058 |
3.822 ± 0.037 | 7.297 ± 0.08 |
2.211 ± 0.031 | 6.597 ± 0.038 |
5.148 ± 0.055 | 10.368 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.498 ± 0.029 | 4.072 ± 0.06 |
4.233 ± 0.038 | 4.031 ± 0.041 |
4.902 ± 0.05 | 6.326 ± 0.046 |
5.555 ± 0.074 | 6.978 ± 0.048 |
1.18 ± 0.023 | 2.886 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
