
Simian torque teno virus 31
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus; Torque teno chlorocebus virus 3
Average proteome isoelectric point is 8.48
Get precalculated fractions of proteins
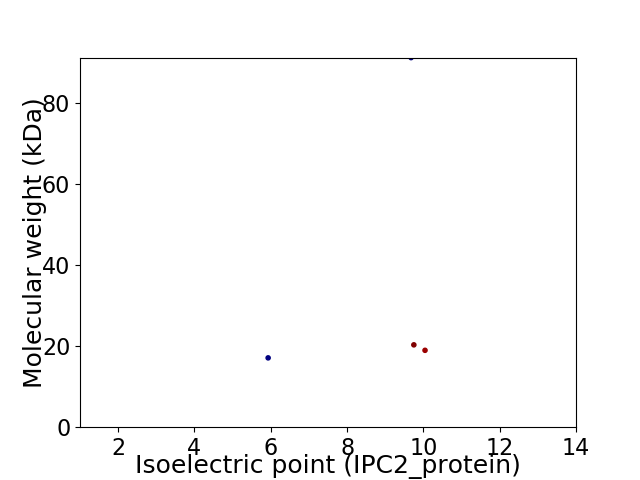
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IMV2|A0A0C5IMV2_9VIRU Capsid protein OS=Simian torque teno virus 31 OX=1619219 PE=3 SV=1
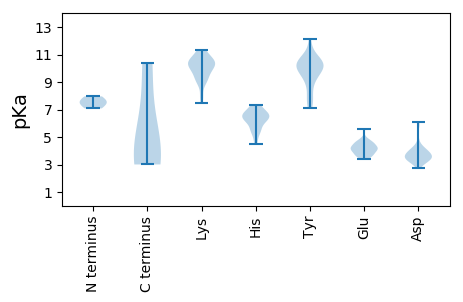
MM1 pKa = 7.08SQLHH5 pKa = 5.06FRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84PRR12 pKa = 11.84PPVHH16 pKa = 6.23GQTGQHH22 pKa = 6.62PSDD25 pKa = 3.75LHH27 pKa = 5.79MFWKK31 pKa = 10.54APQEE35 pKa = 3.93DD36 pKa = 4.02ARR38 pKa = 11.84TLEE41 pKa = 4.28TRR43 pKa = 11.84WLRR46 pKa = 11.84MVLMSHH52 pKa = 6.22STFCHH57 pKa = 6.17CLHH60 pKa = 7.1PLGHH64 pKa = 6.74LHH66 pKa = 6.41YY67 pKa = 10.59ISVRR71 pKa = 11.84DD72 pKa = 4.19KK73 pKa = 11.04GPHH76 pKa = 5.56GQGQTPPRR84 pKa = 11.84TPQLDD89 pKa = 3.86VPPNTPVTTSTPANSTGGSQSLSSGRR115 pKa = 11.84GSWPGTGGEE124 pKa = 4.36EE125 pKa = 3.86EE126 pKa = 4.88GEE128 pKa = 4.08AGGAGAGGIDD138 pKa = 4.84DD139 pKa = 4.14YY140 pKa = 12.09SEE142 pKa = 4.94ADD144 pKa = 3.61LAEE147 pKa = 4.83LYY149 pKa = 10.85AAVDD153 pKa = 3.48GDD155 pKa = 3.78AAGRR159 pKa = 3.68
MM1 pKa = 7.08SQLHH5 pKa = 5.06FRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84PRR12 pKa = 11.84PPVHH16 pKa = 6.23GQTGQHH22 pKa = 6.62PSDD25 pKa = 3.75LHH27 pKa = 5.79MFWKK31 pKa = 10.54APQEE35 pKa = 3.93DD36 pKa = 4.02ARR38 pKa = 11.84TLEE41 pKa = 4.28TRR43 pKa = 11.84WLRR46 pKa = 11.84MVLMSHH52 pKa = 6.22STFCHH57 pKa = 6.17CLHH60 pKa = 7.1PLGHH64 pKa = 6.74LHH66 pKa = 6.41YY67 pKa = 10.59ISVRR71 pKa = 11.84DD72 pKa = 4.19KK73 pKa = 11.04GPHH76 pKa = 5.56GQGQTPPRR84 pKa = 11.84TPQLDD89 pKa = 3.86VPPNTPVTTSTPANSTGGSQSLSSGRR115 pKa = 11.84GSWPGTGGEE124 pKa = 4.36EE125 pKa = 3.86EE126 pKa = 4.88GEE128 pKa = 4.08AGGAGAGGIDD138 pKa = 4.84DD139 pKa = 4.14YY140 pKa = 12.09SEE142 pKa = 4.94ADD144 pKa = 3.61LAEE147 pKa = 4.83LYY149 pKa = 10.85AAVDD153 pKa = 3.48GDD155 pKa = 3.78AAGRR159 pKa = 3.68
Molecular weight: 17.09 kDa
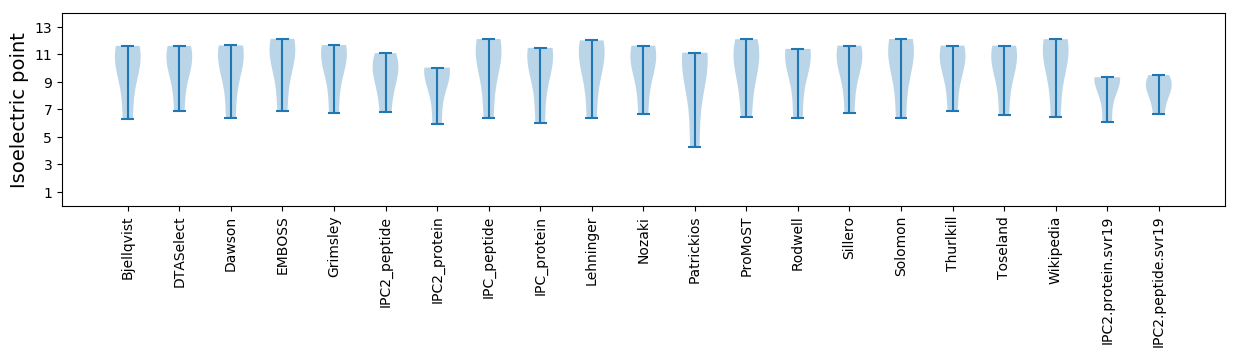
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IMV2|A0A0C5IMV2_9VIRU Capsid protein OS=Simian torque teno virus 31 OX=1619219 PE=3 SV=1
MM1 pKa = 7.45AWYY4 pKa = 8.93GWRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84GWGRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84WRR19 pKa = 11.84YY20 pKa = 9.39RR21 pKa = 11.84RR22 pKa = 11.84LLRR25 pKa = 11.84GRR27 pKa = 11.84PRR29 pKa = 11.84RR30 pKa = 11.84ALRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84GWRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84WQVRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84WGRR50 pKa = 11.84RR51 pKa = 11.84WRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84HH57 pKa = 4.52RR58 pKa = 11.84RR59 pKa = 11.84PRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84YY66 pKa = 7.68RR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84TLALRR79 pKa = 11.84QWNPTTVRR87 pKa = 11.84GCSIRR92 pKa = 11.84GWFPFLITGLGRR104 pKa = 11.84LPFLYY109 pKa = 9.05THH111 pKa = 6.72HH112 pKa = 7.01MDD114 pKa = 6.08DD115 pKa = 3.88IPAEE119 pKa = 4.12KK120 pKa = 9.74MSFGGGLAVSRR131 pKa = 11.84FSLSVLYY138 pKa = 10.37EE139 pKa = 3.85EE140 pKa = 5.59YY141 pKa = 10.17RR142 pKa = 11.84RR143 pKa = 11.84HH144 pKa = 5.37HH145 pKa = 6.22NKK147 pKa = 9.18WSRR150 pKa = 11.84SNVDD154 pKa = 3.69LDD156 pKa = 3.51LLRR159 pKa = 11.84YY160 pKa = 9.68HH161 pKa = 6.76GVSFKK166 pKa = 10.51FYY168 pKa = 10.33RR169 pKa = 11.84EE170 pKa = 3.97PEE172 pKa = 3.43IDD174 pKa = 5.87FIVHH178 pKa = 6.41WSLEE182 pKa = 4.25TPMEE186 pKa = 4.6INLMTHH192 pKa = 6.23MSLHH196 pKa = 6.58PLLLLLNKK204 pKa = 9.84HH205 pKa = 6.37KK206 pKa = 10.59IIIPSMKK213 pKa = 9.15TRR215 pKa = 11.84PHH217 pKa = 5.56GRR219 pKa = 11.84RR220 pKa = 11.84AVRR223 pKa = 11.84LTLPPPRR230 pKa = 11.84LMTNKK235 pKa = 9.54WYY237 pKa = 7.95FTKK240 pKa = 10.57DD241 pKa = 3.07FCKK244 pKa = 10.67VGLCMLMATPCSLLYY259 pKa = 10.06PWLDD263 pKa = 3.47PGKK266 pKa = 9.02TSPCVHH272 pKa = 6.06FHH274 pKa = 7.2ILDD277 pKa = 3.34QSYY280 pKa = 10.51YY281 pKa = 10.6KK282 pKa = 10.2HH283 pKa = 6.61IPNIDD288 pKa = 3.42PNKK291 pKa = 8.01YY292 pKa = 7.46TEE294 pKa = 4.47RR295 pKa = 11.84EE296 pKa = 3.86NDD298 pKa = 3.4LKK300 pKa = 11.16DD301 pKa = 3.13ILKK304 pKa = 9.87IDD306 pKa = 4.36GSSSTCVGACHH317 pKa = 7.36CYY319 pKa = 9.97TEE321 pKa = 4.71LFRR324 pKa = 11.84DD325 pKa = 3.76AWPSIRR331 pKa = 11.84KK332 pKa = 8.98PGVASQPFGLTEE344 pKa = 3.58ICKK347 pKa = 7.92TQNYY351 pKa = 7.11TNNSLADD358 pKa = 3.74TFISKK363 pKa = 10.72ANEE366 pKa = 3.5AHH368 pKa = 7.36SNITDD373 pKa = 3.37KK374 pKa = 11.32LMPCVYY380 pKa = 9.85GSNSAVITQLKK391 pKa = 9.33SSKK394 pKa = 10.61DD395 pKa = 3.17KK396 pKa = 11.09TFMHH400 pKa = 6.59RR401 pKa = 11.84WGLYY405 pKa = 7.3SHH407 pKa = 6.85YY408 pKa = 10.53FLTTRR413 pKa = 11.84RR414 pKa = 11.84LDD416 pKa = 3.67PQLTGPTIKK425 pKa = 10.41VGYY428 pKa = 9.63NPQTDD433 pKa = 3.55EE434 pKa = 4.48GKK436 pKa = 11.14GNLIFLQPLTRR447 pKa = 11.84SDD449 pKa = 3.85TIFNPTSSKK458 pKa = 10.6CAIFDD463 pKa = 3.86TPLWLALYY471 pKa = 9.85GYY473 pKa = 10.23EE474 pKa = 4.26SYY476 pKa = 9.35CTKK479 pKa = 10.46FLNDD483 pKa = 3.37EE484 pKa = 4.12QVMYY488 pKa = 10.95NYY490 pKa = 9.56VLCVRR495 pKa = 11.84CTYY498 pKa = 8.11TWPRR502 pKa = 11.84MYY504 pKa = 10.05RR505 pKa = 11.84TGNPNQCDD513 pKa = 3.36IPLGNSFLEE522 pKa = 4.61GNMPGGAYY530 pKa = 8.45PPPLLMRR537 pKa = 11.84TKK539 pKa = 9.86WYY541 pKa = 8.98PCLLHH546 pKa = 6.78QRR548 pKa = 11.84EE549 pKa = 4.62FINAVVGSGPFMPRR563 pKa = 11.84GYY565 pKa = 10.09SVKK568 pKa = 10.05SWEE571 pKa = 3.89LTAGYY576 pKa = 10.21KK577 pKa = 10.18FRR579 pKa = 11.84FSWGGHH585 pKa = 4.7APYY588 pKa = 9.57TKK590 pKa = 10.3QVDD593 pKa = 4.06DD594 pKa = 3.96PCKK597 pKa = 10.31KK598 pKa = 9.13GTHH601 pKa = 6.61ALPDD605 pKa = 3.98PDD607 pKa = 3.88RR608 pKa = 11.84QRR610 pKa = 11.84MAVQVEE616 pKa = 4.33DD617 pKa = 4.46PGRR620 pKa = 11.84LDD622 pKa = 3.38PQYY625 pKa = 10.33MFHH628 pKa = 6.74GWDD631 pKa = 3.07QRR633 pKa = 11.84RR634 pKa = 11.84GYY636 pKa = 10.37ILGRR640 pKa = 11.84AIKK643 pKa = 10.01RR644 pKa = 11.84VSEE647 pKa = 4.23DD648 pKa = 3.53PGHH651 pKa = 7.3DD652 pKa = 3.34SDD654 pKa = 5.92IPTGPLKK661 pKa = 10.48RR662 pKa = 11.84PRR664 pKa = 11.84VDD666 pKa = 3.58APPEE670 pKa = 4.46GGWPDD675 pKa = 4.08PEE677 pKa = 4.68RR678 pKa = 11.84GWISSLLQEE687 pKa = 4.46TPPGRR692 pKa = 11.84RR693 pKa = 11.84HH694 pKa = 6.16SPPLQLQLPRR704 pKa = 11.84EE705 pKa = 4.3PEE707 pKa = 3.62EE708 pKa = 3.73TTRR711 pKa = 11.84RR712 pKa = 11.84RR713 pKa = 11.84RR714 pKa = 11.84RR715 pKa = 11.84SEE717 pKa = 4.18GEE719 pKa = 3.61IPPPEE724 pKa = 3.74TAAASTQAFLEE735 pKa = 4.51LEE737 pKa = 4.53LEE739 pKa = 4.26RR740 pKa = 11.84QRR742 pKa = 11.84QQQHH746 pKa = 5.67RR747 pKa = 11.84LRR749 pKa = 11.84KK750 pKa = 9.21GLQDD754 pKa = 3.58LFSRR758 pKa = 11.84LVRR761 pKa = 11.84TEE763 pKa = 4.22GGHH766 pKa = 5.42QVDD769 pKa = 4.08PRR771 pKa = 11.84LLL773 pKa = 4.28
MM1 pKa = 7.45AWYY4 pKa = 8.93GWRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84GWGRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84WRR19 pKa = 11.84YY20 pKa = 9.39RR21 pKa = 11.84RR22 pKa = 11.84LLRR25 pKa = 11.84GRR27 pKa = 11.84PRR29 pKa = 11.84RR30 pKa = 11.84ALRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84GWRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84WQVRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84WGRR50 pKa = 11.84RR51 pKa = 11.84WRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84HH57 pKa = 4.52RR58 pKa = 11.84RR59 pKa = 11.84PRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84YY66 pKa = 7.68RR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84TLALRR79 pKa = 11.84QWNPTTVRR87 pKa = 11.84GCSIRR92 pKa = 11.84GWFPFLITGLGRR104 pKa = 11.84LPFLYY109 pKa = 9.05THH111 pKa = 6.72HH112 pKa = 7.01MDD114 pKa = 6.08DD115 pKa = 3.88IPAEE119 pKa = 4.12KK120 pKa = 9.74MSFGGGLAVSRR131 pKa = 11.84FSLSVLYY138 pKa = 10.37EE139 pKa = 3.85EE140 pKa = 5.59YY141 pKa = 10.17RR142 pKa = 11.84RR143 pKa = 11.84HH144 pKa = 5.37HH145 pKa = 6.22NKK147 pKa = 9.18WSRR150 pKa = 11.84SNVDD154 pKa = 3.69LDD156 pKa = 3.51LLRR159 pKa = 11.84YY160 pKa = 9.68HH161 pKa = 6.76GVSFKK166 pKa = 10.51FYY168 pKa = 10.33RR169 pKa = 11.84EE170 pKa = 3.97PEE172 pKa = 3.43IDD174 pKa = 5.87FIVHH178 pKa = 6.41WSLEE182 pKa = 4.25TPMEE186 pKa = 4.6INLMTHH192 pKa = 6.23MSLHH196 pKa = 6.58PLLLLLNKK204 pKa = 9.84HH205 pKa = 6.37KK206 pKa = 10.59IIIPSMKK213 pKa = 9.15TRR215 pKa = 11.84PHH217 pKa = 5.56GRR219 pKa = 11.84RR220 pKa = 11.84AVRR223 pKa = 11.84LTLPPPRR230 pKa = 11.84LMTNKK235 pKa = 9.54WYY237 pKa = 7.95FTKK240 pKa = 10.57DD241 pKa = 3.07FCKK244 pKa = 10.67VGLCMLMATPCSLLYY259 pKa = 10.06PWLDD263 pKa = 3.47PGKK266 pKa = 9.02TSPCVHH272 pKa = 6.06FHH274 pKa = 7.2ILDD277 pKa = 3.34QSYY280 pKa = 10.51YY281 pKa = 10.6KK282 pKa = 10.2HH283 pKa = 6.61IPNIDD288 pKa = 3.42PNKK291 pKa = 8.01YY292 pKa = 7.46TEE294 pKa = 4.47RR295 pKa = 11.84EE296 pKa = 3.86NDD298 pKa = 3.4LKK300 pKa = 11.16DD301 pKa = 3.13ILKK304 pKa = 9.87IDD306 pKa = 4.36GSSSTCVGACHH317 pKa = 7.36CYY319 pKa = 9.97TEE321 pKa = 4.71LFRR324 pKa = 11.84DD325 pKa = 3.76AWPSIRR331 pKa = 11.84KK332 pKa = 8.98PGVASQPFGLTEE344 pKa = 3.58ICKK347 pKa = 7.92TQNYY351 pKa = 7.11TNNSLADD358 pKa = 3.74TFISKK363 pKa = 10.72ANEE366 pKa = 3.5AHH368 pKa = 7.36SNITDD373 pKa = 3.37KK374 pKa = 11.32LMPCVYY380 pKa = 9.85GSNSAVITQLKK391 pKa = 9.33SSKK394 pKa = 10.61DD395 pKa = 3.17KK396 pKa = 11.09TFMHH400 pKa = 6.59RR401 pKa = 11.84WGLYY405 pKa = 7.3SHH407 pKa = 6.85YY408 pKa = 10.53FLTTRR413 pKa = 11.84RR414 pKa = 11.84LDD416 pKa = 3.67PQLTGPTIKK425 pKa = 10.41VGYY428 pKa = 9.63NPQTDD433 pKa = 3.55EE434 pKa = 4.48GKK436 pKa = 11.14GNLIFLQPLTRR447 pKa = 11.84SDD449 pKa = 3.85TIFNPTSSKK458 pKa = 10.6CAIFDD463 pKa = 3.86TPLWLALYY471 pKa = 9.85GYY473 pKa = 10.23EE474 pKa = 4.26SYY476 pKa = 9.35CTKK479 pKa = 10.46FLNDD483 pKa = 3.37EE484 pKa = 4.12QVMYY488 pKa = 10.95NYY490 pKa = 9.56VLCVRR495 pKa = 11.84CTYY498 pKa = 8.11TWPRR502 pKa = 11.84MYY504 pKa = 10.05RR505 pKa = 11.84TGNPNQCDD513 pKa = 3.36IPLGNSFLEE522 pKa = 4.61GNMPGGAYY530 pKa = 8.45PPPLLMRR537 pKa = 11.84TKK539 pKa = 9.86WYY541 pKa = 8.98PCLLHH546 pKa = 6.78QRR548 pKa = 11.84EE549 pKa = 4.62FINAVVGSGPFMPRR563 pKa = 11.84GYY565 pKa = 10.09SVKK568 pKa = 10.05SWEE571 pKa = 3.89LTAGYY576 pKa = 10.21KK577 pKa = 10.18FRR579 pKa = 11.84FSWGGHH585 pKa = 4.7APYY588 pKa = 9.57TKK590 pKa = 10.3QVDD593 pKa = 4.06DD594 pKa = 3.96PCKK597 pKa = 10.31KK598 pKa = 9.13GTHH601 pKa = 6.61ALPDD605 pKa = 3.98PDD607 pKa = 3.88RR608 pKa = 11.84QRR610 pKa = 11.84MAVQVEE616 pKa = 4.33DD617 pKa = 4.46PGRR620 pKa = 11.84LDD622 pKa = 3.38PQYY625 pKa = 10.33MFHH628 pKa = 6.74GWDD631 pKa = 3.07QRR633 pKa = 11.84RR634 pKa = 11.84GYY636 pKa = 10.37ILGRR640 pKa = 11.84AIKK643 pKa = 10.01RR644 pKa = 11.84VSEE647 pKa = 4.23DD648 pKa = 3.53PGHH651 pKa = 7.3DD652 pKa = 3.34SDD654 pKa = 5.92IPTGPLKK661 pKa = 10.48RR662 pKa = 11.84PRR664 pKa = 11.84VDD666 pKa = 3.58APPEE670 pKa = 4.46GGWPDD675 pKa = 4.08PEE677 pKa = 4.68RR678 pKa = 11.84GWISSLLQEE687 pKa = 4.46TPPGRR692 pKa = 11.84RR693 pKa = 11.84HH694 pKa = 6.16SPPLQLQLPRR704 pKa = 11.84EE705 pKa = 4.3PEE707 pKa = 3.62EE708 pKa = 3.73TTRR711 pKa = 11.84RR712 pKa = 11.84RR713 pKa = 11.84RR714 pKa = 11.84RR715 pKa = 11.84SEE717 pKa = 4.18GEE719 pKa = 3.61IPPPEE724 pKa = 3.74TAAASTQAFLEE735 pKa = 4.51LEE737 pKa = 4.53LEE739 pKa = 4.26RR740 pKa = 11.84QRR742 pKa = 11.84QQQHH746 pKa = 5.67RR747 pKa = 11.84LRR749 pKa = 11.84KK750 pKa = 9.21GLQDD754 pKa = 3.58LFSRR758 pKa = 11.84LVRR761 pKa = 11.84TEE763 pKa = 4.22GGHH766 pKa = 5.42QVDD769 pKa = 4.08PRR771 pKa = 11.84LLL773 pKa = 4.28
Molecular weight: 91.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1289 |
159 |
773 |
322.3 |
36.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.896 ± 2.389 | 1.629 ± 0.501 |
4.81 ± 0.248 | 4.344 ± 0.144 |
3.181 ± 0.46 | 8.301 ± 1.36 |
3.103 ± 0.833 | 3.336 ± 0.57 |
3.724 ± 0.788 | 7.603 ± 2.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.095 ± 0.383 | 2.56 ± 0.615 |
9.465 ± 1.805 | 4.112 ± 0.555 |
12.645 ± 1.244 | 7.448 ± 2.382 |
6.129 ± 0.41 | 3.646 ± 0.493 |
2.638 ± 0.755 | 3.336 ± 0.797 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
