
Betacoronavirus Erinaceus/VMC/DEU/2012
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Merbecovirus; Hedgehog coronavirus 1
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
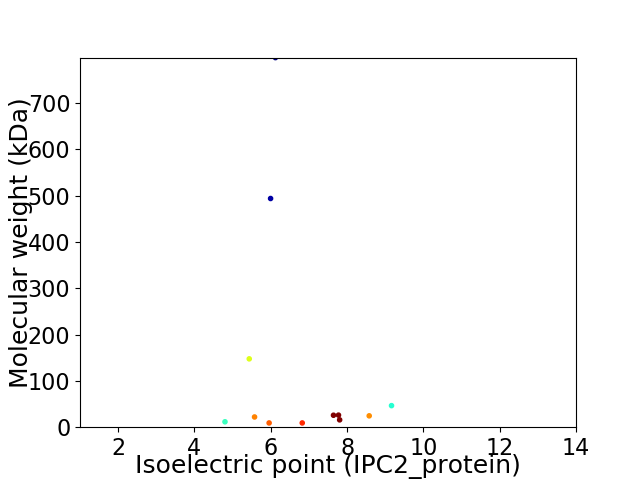
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
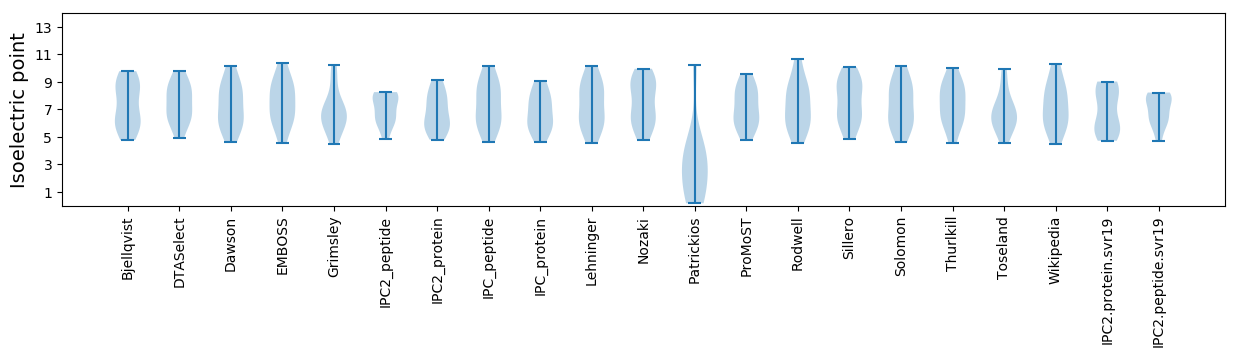
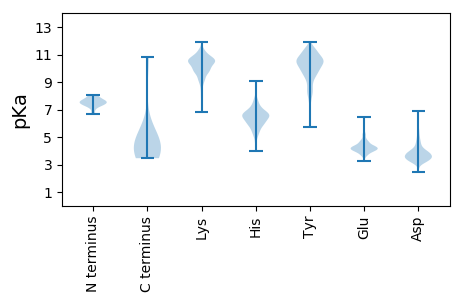
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5LMN3|U5LMN3_9BETC Orf5 OS=Betacoronavirus Erinaceus/VMC/DEU/2012 OX=1385427 GN=orf5 PE=4 SV=1
MM1 pKa = 6.7QSKK4 pKa = 10.5RR5 pKa = 11.84GILTLIVCNILAMSLAKK22 pKa = 10.44HH23 pKa = 6.69FIPEE27 pKa = 3.56HH28 pKa = 5.26CANYY32 pKa = 9.38EE33 pKa = 3.68GAMFHH38 pKa = 7.09ACVQNAISTAAGTYY52 pKa = 8.98TNTKK56 pKa = 9.65LSYY59 pKa = 10.29PVFDD63 pKa = 4.2NDD65 pKa = 3.6GVTYY69 pKa = 11.05YY70 pKa = 10.96DD71 pKa = 3.3ATQDD75 pKa = 3.27RR76 pKa = 11.84DD77 pKa = 3.51ATPSYY82 pKa = 9.78EE83 pKa = 3.66HH84 pKa = 7.15SEE86 pKa = 4.33FYY88 pKa = 10.94EE89 pKa = 4.25LDD91 pKa = 3.5EE92 pKa = 5.16SSFSAKK98 pKa = 9.6PVLAEE103 pKa = 4.03LQQ105 pKa = 3.46
MM1 pKa = 6.7QSKK4 pKa = 10.5RR5 pKa = 11.84GILTLIVCNILAMSLAKK22 pKa = 10.44HH23 pKa = 6.69FIPEE27 pKa = 3.56HH28 pKa = 5.26CANYY32 pKa = 9.38EE33 pKa = 3.68GAMFHH38 pKa = 7.09ACVQNAISTAAGTYY52 pKa = 8.98TNTKK56 pKa = 9.65LSYY59 pKa = 10.29PVFDD63 pKa = 4.2NDD65 pKa = 3.6GVTYY69 pKa = 11.05YY70 pKa = 10.96DD71 pKa = 3.3ATQDD75 pKa = 3.27RR76 pKa = 11.84DD77 pKa = 3.51ATPSYY82 pKa = 9.78EE83 pKa = 3.66HH84 pKa = 7.15SEE86 pKa = 4.33FYY88 pKa = 10.94EE89 pKa = 4.25LDD91 pKa = 3.5EE92 pKa = 5.16SSFSAKK98 pKa = 9.6PVLAEE103 pKa = 4.03LQQ105 pKa = 3.46
Molecular weight: 11.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5LR11|U5LR11_9BETC 3C-like proteinase OS=Betacoronavirus Erinaceus/VMC/DEU/2012 OX=1385427 GN=ORF 1a PE=3 SV=1
MM1 pKa = 7.64ATPQQPRR8 pKa = 11.84AVTFADD14 pKa = 3.85NNGNQQNGNNRR25 pKa = 11.84GRR27 pKa = 11.84PRR29 pKa = 11.84QPKK32 pKa = 8.87PRR34 pKa = 11.84PAPNVSVSWYY44 pKa = 9.74TGITQHH50 pKa = 6.06GKK52 pKa = 9.7QPLAFPAGQGVPLNANSTPKK72 pKa = 10.38QNAGYY77 pKa = 9.38WRR79 pKa = 11.84RR80 pKa = 11.84QDD82 pKa = 3.65RR83 pKa = 11.84KK84 pKa = 10.35LNTGNGVKK92 pKa = 9.99QLAPRR97 pKa = 11.84WYY99 pKa = 9.84FYY101 pKa = 10.45YY102 pKa = 10.42TGTGPEE108 pKa = 3.91ANLPFRR114 pKa = 11.84TVKK117 pKa = 10.79DD118 pKa = 3.7GIYY121 pKa = 9.43WVWEE125 pKa = 4.34EE126 pKa = 4.44GASEE130 pKa = 4.17APSDD134 pKa = 3.72FGTRR138 pKa = 11.84NPANDD143 pKa = 3.63AAIVTQFAPGTQLPKK158 pKa = 9.78NCHH161 pKa = 5.54IEE163 pKa = 4.21GTGGNSQSSSRR174 pKa = 11.84ASSASRR180 pKa = 11.84NSSRR184 pKa = 11.84SNSRR188 pKa = 11.84GSQPGSRR195 pKa = 11.84SHH197 pKa = 6.83SPGSVGPSDD206 pKa = 4.68ASALLYY212 pKa = 11.02LEE214 pKa = 4.08LAKK217 pKa = 10.7RR218 pKa = 11.84LEE220 pKa = 3.99ALEE223 pKa = 4.35AGKK226 pKa = 10.58SKK228 pKa = 10.59SAPKK232 pKa = 10.61VITKK236 pKa = 10.2KK237 pKa = 10.55DD238 pKa = 3.07AADD241 pKa = 3.38AKK243 pKa = 10.87KK244 pKa = 10.6KK245 pKa = 9.11MRR247 pKa = 11.84HH248 pKa = 5.25KK249 pKa = 10.55RR250 pKa = 11.84VATKK254 pKa = 10.31AYY256 pKa = 10.64NPTQAFGLRR265 pKa = 11.84GPGDD269 pKa = 3.51LQGNFGDD276 pKa = 5.16LKK278 pKa = 9.27YY279 pKa = 10.76CKK281 pKa = 9.97EE282 pKa = 4.28GVDD285 pKa = 4.07DD286 pKa = 4.43PRR288 pKa = 11.84WPQMAEE294 pKa = 3.91LAPSASAFLSMSQLKK309 pKa = 9.78LVHH312 pKa = 6.65HH313 pKa = 7.09SNDD316 pKa = 3.31TDD318 pKa = 3.53NKK320 pKa = 7.67PVYY323 pKa = 8.78MLRR326 pKa = 11.84YY327 pKa = 9.45SGAIKK332 pKa = 10.61LDD334 pKa = 3.7PKK336 pKa = 10.55NPNYY340 pKa = 10.73SKK342 pKa = 10.29WLEE345 pKa = 3.92ILEE348 pKa = 4.42GNIDD352 pKa = 3.55AYY354 pKa = 10.34KK355 pKa = 10.71NFPKK359 pKa = 10.26KK360 pKa = 9.87EE361 pKa = 4.02KK362 pKa = 9.93KK363 pKa = 9.84QKK365 pKa = 9.97QSQIKK370 pKa = 9.79QEE372 pKa = 4.21AEE374 pKa = 3.8DD375 pKa = 4.7PEE377 pKa = 4.57WDD379 pKa = 3.7STDD382 pKa = 4.88LFSQPLQDD390 pKa = 5.14PDD392 pKa = 3.92NQPKK396 pKa = 9.44AQRR399 pKa = 11.84VPKK402 pKa = 10.63GSITQRR408 pKa = 11.84SRR410 pKa = 11.84PPKK413 pKa = 7.77PTVGPMVDD421 pKa = 3.43VEE423 pKa = 4.21PP424 pKa = 4.76
MM1 pKa = 7.64ATPQQPRR8 pKa = 11.84AVTFADD14 pKa = 3.85NNGNQQNGNNRR25 pKa = 11.84GRR27 pKa = 11.84PRR29 pKa = 11.84QPKK32 pKa = 8.87PRR34 pKa = 11.84PAPNVSVSWYY44 pKa = 9.74TGITQHH50 pKa = 6.06GKK52 pKa = 9.7QPLAFPAGQGVPLNANSTPKK72 pKa = 10.38QNAGYY77 pKa = 9.38WRR79 pKa = 11.84RR80 pKa = 11.84QDD82 pKa = 3.65RR83 pKa = 11.84KK84 pKa = 10.35LNTGNGVKK92 pKa = 9.99QLAPRR97 pKa = 11.84WYY99 pKa = 9.84FYY101 pKa = 10.45YY102 pKa = 10.42TGTGPEE108 pKa = 3.91ANLPFRR114 pKa = 11.84TVKK117 pKa = 10.79DD118 pKa = 3.7GIYY121 pKa = 9.43WVWEE125 pKa = 4.34EE126 pKa = 4.44GASEE130 pKa = 4.17APSDD134 pKa = 3.72FGTRR138 pKa = 11.84NPANDD143 pKa = 3.63AAIVTQFAPGTQLPKK158 pKa = 9.78NCHH161 pKa = 5.54IEE163 pKa = 4.21GTGGNSQSSSRR174 pKa = 11.84ASSASRR180 pKa = 11.84NSSRR184 pKa = 11.84SNSRR188 pKa = 11.84GSQPGSRR195 pKa = 11.84SHH197 pKa = 6.83SPGSVGPSDD206 pKa = 4.68ASALLYY212 pKa = 11.02LEE214 pKa = 4.08LAKK217 pKa = 10.7RR218 pKa = 11.84LEE220 pKa = 3.99ALEE223 pKa = 4.35AGKK226 pKa = 10.58SKK228 pKa = 10.59SAPKK232 pKa = 10.61VITKK236 pKa = 10.2KK237 pKa = 10.55DD238 pKa = 3.07AADD241 pKa = 3.38AKK243 pKa = 10.87KK244 pKa = 10.6KK245 pKa = 9.11MRR247 pKa = 11.84HH248 pKa = 5.25KK249 pKa = 10.55RR250 pKa = 11.84VATKK254 pKa = 10.31AYY256 pKa = 10.64NPTQAFGLRR265 pKa = 11.84GPGDD269 pKa = 3.51LQGNFGDD276 pKa = 5.16LKK278 pKa = 9.27YY279 pKa = 10.76CKK281 pKa = 9.97EE282 pKa = 4.28GVDD285 pKa = 4.07DD286 pKa = 4.43PRR288 pKa = 11.84WPQMAEE294 pKa = 3.91LAPSASAFLSMSQLKK309 pKa = 9.78LVHH312 pKa = 6.65HH313 pKa = 7.09SNDD316 pKa = 3.31TDD318 pKa = 3.53NKK320 pKa = 7.67PVYY323 pKa = 8.78MLRR326 pKa = 11.84YY327 pKa = 9.45SGAIKK332 pKa = 10.61LDD334 pKa = 3.7PKK336 pKa = 10.55NPNYY340 pKa = 10.73SKK342 pKa = 10.29WLEE345 pKa = 3.92ILEE348 pKa = 4.42GNIDD352 pKa = 3.55AYY354 pKa = 10.34KK355 pKa = 10.71NFPKK359 pKa = 10.26KK360 pKa = 9.87EE361 pKa = 4.02KK362 pKa = 9.93KK363 pKa = 9.84QKK365 pKa = 9.97QSQIKK370 pKa = 9.79QEE372 pKa = 4.21AEE374 pKa = 3.8DD375 pKa = 4.7PEE377 pKa = 4.57WDD379 pKa = 3.7STDD382 pKa = 4.88LFSQPLQDD390 pKa = 5.14PDD392 pKa = 3.92NQPKK396 pKa = 9.44AQRR399 pKa = 11.84VPKK402 pKa = 10.63GSITQRR408 pKa = 11.84SRR410 pKa = 11.84PPKK413 pKa = 7.77PTVGPMVDD421 pKa = 3.43VEE423 pKa = 4.21PP424 pKa = 4.76
Molecular weight: 46.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14639 |
81 |
7150 |
1219.9 |
135.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.022 ± 0.198 | 3.224 ± 0.254 |
5.103 ± 0.374 | 4.317 ± 0.303 |
4.871 ± 0.18 | 5.718 ± 0.444 |
2.008 ± 0.156 | 4.509 ± 0.357 |
5.929 ± 0.453 | 9.584 ± 0.521 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.527 ± 0.214 | 5.595 ± 0.252 |
3.907 ± 0.58 | 3.696 ± 0.495 |
3.156 ± 0.255 | 7.576 ± 0.398 |
5.991 ± 0.282 | 9.488 ± 0.807 |
1.141 ± 0.12 | 4.638 ± 0.284 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
