
Bacillus oleivorans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
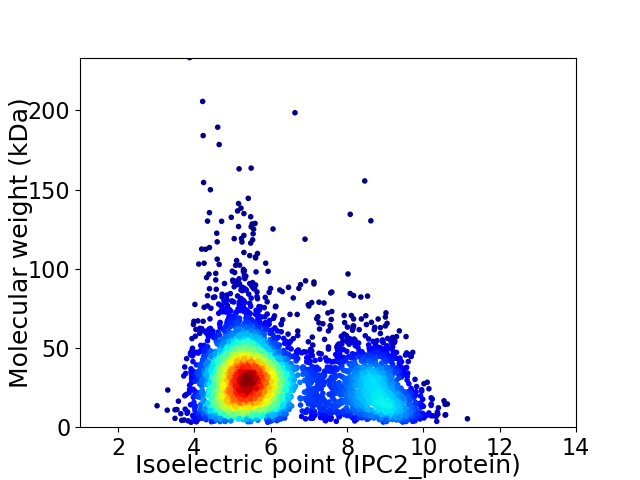
Virtual 2D-PAGE plot for 4443 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
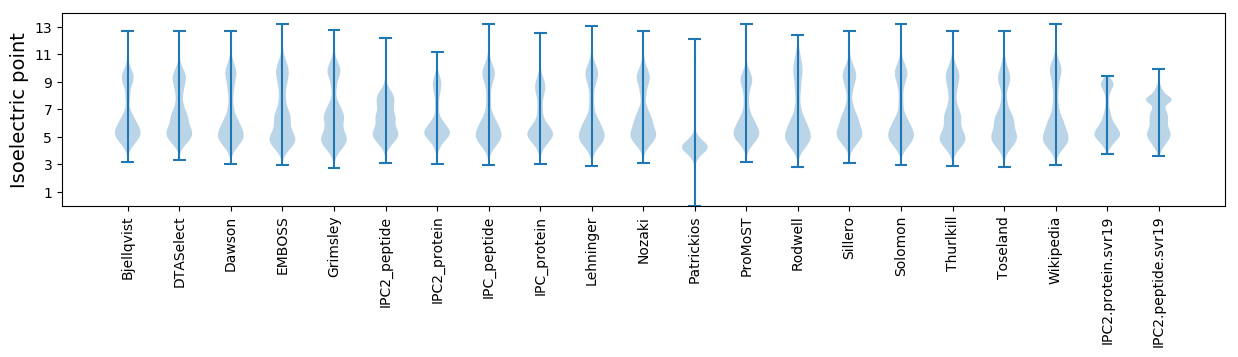
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A285CH50|A0A285CH50_9BACI Cardiolipin synthase OS=Bacillus oleivorans OX=1448271 GN=SAMN05877753_101240 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.44KK3 pKa = 9.65NWLIKK8 pKa = 10.46GFALLLIMMLLVAGCSQDD26 pKa = 3.71ADD28 pKa = 3.53EE29 pKa = 5.36GQGNTNEE36 pKa = 4.25PDD38 pKa = 3.35SGEE41 pKa = 4.27TTDD44 pKa = 5.17DD45 pKa = 3.38QDD47 pKa = 3.76NGTTEE52 pKa = 4.22EE53 pKa = 4.17PAEE56 pKa = 4.09PLTVKK61 pKa = 10.39FAAQNDD67 pKa = 3.99NTPATQAAIDD77 pKa = 4.36AFNASQDD84 pKa = 3.54QYY86 pKa = 9.09VVEE89 pKa = 4.25WVEE92 pKa = 3.95MTNDD96 pKa = 3.23SGQMHH101 pKa = 7.06DD102 pKa = 3.72QLLNSLSSGSSEE114 pKa = 4.1YY115 pKa = 10.88DD116 pKa = 3.41VLSLDD121 pKa = 3.99VVWAGEE127 pKa = 4.02FAGAGYY133 pKa = 10.54LEE135 pKa = 5.27PIDD138 pKa = 4.42VKK140 pKa = 10.35MDD142 pKa = 3.4EE143 pKa = 4.69AGYY146 pKa = 10.74SIDD149 pKa = 4.55DD150 pKa = 3.97FNAGSMTSGNYY161 pKa = 9.87KK162 pKa = 10.31GIQYY166 pKa = 7.42TLPFFPDD173 pKa = 3.49LGLLYY178 pKa = 10.17YY179 pKa = 10.6RR180 pKa = 11.84SDD182 pKa = 3.06IVSEE186 pKa = 3.72EE187 pKa = 4.04DD188 pKa = 3.26AAKK191 pKa = 10.65LEE193 pKa = 4.44SGEE196 pKa = 4.05YY197 pKa = 9.46TYY199 pKa = 11.54EE200 pKa = 3.99EE201 pKa = 4.65LGQMAEE207 pKa = 4.12KK208 pKa = 10.71YY209 pKa = 8.63MNEE212 pKa = 4.0SGTSYY217 pKa = 11.33GFVFQSAQYY226 pKa = 10.7EE227 pKa = 4.5GLTVNATEE235 pKa = 4.38FTSEE239 pKa = 4.23YY240 pKa = 10.12KK241 pKa = 10.61DD242 pKa = 3.54LSAGLEE248 pKa = 4.07TMYY251 pKa = 11.28SFVTSSFVPTDD262 pKa = 3.01ILNFQEE268 pKa = 5.85GEE270 pKa = 4.04THH272 pKa = 6.9TNFEE276 pKa = 3.69QGNAVFARR284 pKa = 11.84NWPYY288 pKa = 10.61QYY290 pKa = 11.4GRR292 pKa = 11.84IKK294 pKa = 10.48GQEE297 pKa = 4.18DD298 pKa = 4.84GVTVTPEE305 pKa = 3.82QVGIAPLPNGGSVGGWLLSINKK327 pKa = 9.79SSEE330 pKa = 4.13NKK332 pKa = 9.88DD333 pKa = 3.56GAWEE337 pKa = 4.63FIKK340 pKa = 10.75FLAGPEE346 pKa = 4.26GQKK349 pKa = 10.38IMSTGGGYY357 pKa = 10.57LPGYY361 pKa = 9.5NALLEE366 pKa = 4.51DD367 pKa = 4.31EE368 pKa = 4.96EE369 pKa = 5.04VVASNEE375 pKa = 3.99LLSNPGFQNALATTIARR392 pKa = 11.84PVSPDD397 pKa = 3.24YY398 pKa = 11.56AKK400 pKa = 10.96VSDD403 pKa = 4.3TIQVNVHH410 pKa = 6.54KK411 pKa = 10.01YY412 pKa = 9.19LSSGEE417 pKa = 4.09GLEE420 pKa = 4.28EE421 pKa = 3.67AVQAIEE427 pKa = 3.89AALAAEE433 pKa = 4.35
MM1 pKa = 7.47KK2 pKa = 10.44KK3 pKa = 9.65NWLIKK8 pKa = 10.46GFALLLIMMLLVAGCSQDD26 pKa = 3.71ADD28 pKa = 3.53EE29 pKa = 5.36GQGNTNEE36 pKa = 4.25PDD38 pKa = 3.35SGEE41 pKa = 4.27TTDD44 pKa = 5.17DD45 pKa = 3.38QDD47 pKa = 3.76NGTTEE52 pKa = 4.22EE53 pKa = 4.17PAEE56 pKa = 4.09PLTVKK61 pKa = 10.39FAAQNDD67 pKa = 3.99NTPATQAAIDD77 pKa = 4.36AFNASQDD84 pKa = 3.54QYY86 pKa = 9.09VVEE89 pKa = 4.25WVEE92 pKa = 3.95MTNDD96 pKa = 3.23SGQMHH101 pKa = 7.06DD102 pKa = 3.72QLLNSLSSGSSEE114 pKa = 4.1YY115 pKa = 10.88DD116 pKa = 3.41VLSLDD121 pKa = 3.99VVWAGEE127 pKa = 4.02FAGAGYY133 pKa = 10.54LEE135 pKa = 5.27PIDD138 pKa = 4.42VKK140 pKa = 10.35MDD142 pKa = 3.4EE143 pKa = 4.69AGYY146 pKa = 10.74SIDD149 pKa = 4.55DD150 pKa = 3.97FNAGSMTSGNYY161 pKa = 9.87KK162 pKa = 10.31GIQYY166 pKa = 7.42TLPFFPDD173 pKa = 3.49LGLLYY178 pKa = 10.17YY179 pKa = 10.6RR180 pKa = 11.84SDD182 pKa = 3.06IVSEE186 pKa = 3.72EE187 pKa = 4.04DD188 pKa = 3.26AAKK191 pKa = 10.65LEE193 pKa = 4.44SGEE196 pKa = 4.05YY197 pKa = 9.46TYY199 pKa = 11.54EE200 pKa = 3.99EE201 pKa = 4.65LGQMAEE207 pKa = 4.12KK208 pKa = 10.71YY209 pKa = 8.63MNEE212 pKa = 4.0SGTSYY217 pKa = 11.33GFVFQSAQYY226 pKa = 10.7EE227 pKa = 4.5GLTVNATEE235 pKa = 4.38FTSEE239 pKa = 4.23YY240 pKa = 10.12KK241 pKa = 10.61DD242 pKa = 3.54LSAGLEE248 pKa = 4.07TMYY251 pKa = 11.28SFVTSSFVPTDD262 pKa = 3.01ILNFQEE268 pKa = 5.85GEE270 pKa = 4.04THH272 pKa = 6.9TNFEE276 pKa = 3.69QGNAVFARR284 pKa = 11.84NWPYY288 pKa = 10.61QYY290 pKa = 11.4GRR292 pKa = 11.84IKK294 pKa = 10.48GQEE297 pKa = 4.18DD298 pKa = 4.84GVTVTPEE305 pKa = 3.82QVGIAPLPNGGSVGGWLLSINKK327 pKa = 9.79SSEE330 pKa = 4.13NKK332 pKa = 9.88DD333 pKa = 3.56GAWEE337 pKa = 4.63FIKK340 pKa = 10.75FLAGPEE346 pKa = 4.26GQKK349 pKa = 10.38IMSTGGGYY357 pKa = 10.57LPGYY361 pKa = 9.5NALLEE366 pKa = 4.51DD367 pKa = 4.31EE368 pKa = 4.96EE369 pKa = 5.04VVASNEE375 pKa = 3.99LLSNPGFQNALATTIARR392 pKa = 11.84PVSPDD397 pKa = 3.24YY398 pKa = 11.56AKK400 pKa = 10.96VSDD403 pKa = 4.3TIQVNVHH410 pKa = 6.54KK411 pKa = 10.01YY412 pKa = 9.19LSSGEE417 pKa = 4.09GLEE420 pKa = 4.28EE421 pKa = 3.67AVQAIEE427 pKa = 3.89AALAAEE433 pKa = 4.35
Molecular weight: 46.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A285CGU1|A0A285CGU1_9BACI Uncharacterized protein OS=Bacillus oleivorans OX=1448271 GN=SAMN05877753_101129 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.34QPNKK9 pKa = 8.21RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84SKK14 pKa = 10.24VHH16 pKa = 6.26GFRR19 pKa = 11.84QRR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.48VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.94VLSAA44 pKa = 4.11
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.34QPNKK9 pKa = 8.21RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84SKK14 pKa = 10.24VHH16 pKa = 6.26GFRR19 pKa = 11.84QRR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.48VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.94VLSAA44 pKa = 4.11
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1266437 |
29 |
2147 |
285.0 |
32.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.968 ± 0.041 | 0.685 ± 0.012 |
4.981 ± 0.029 | 7.616 ± 0.037 |
4.662 ± 0.034 | 6.962 ± 0.035 |
2.077 ± 0.017 | 8.025 ± 0.037 |
6.629 ± 0.038 | 9.876 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.617 ± 0.018 | 4.327 ± 0.024 |
3.815 ± 0.023 | 3.829 ± 0.022 |
4.017 ± 0.028 | 5.969 ± 0.029 |
5.337 ± 0.025 | 6.971 ± 0.03 |
1.094 ± 0.016 | 3.543 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
