
Phasey bean mild yellows virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 8.26
Get precalculated fractions of proteins
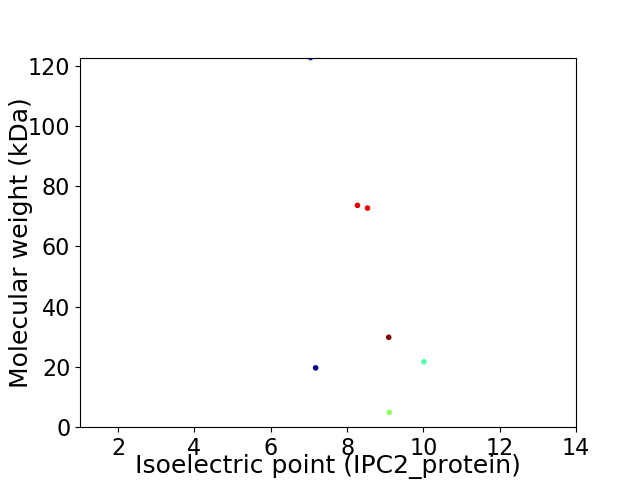
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0S3JNY5|A0A0S3JNY5_9LUTE Movement protein OS=Phasey bean mild yellows virus OX=1756832 GN=MP PE=3 SV=1
MM1 pKa = 7.53NSFSVLFFFFCAFFCVAPVISGPGPVQPEE30 pKa = 3.76WAASKK35 pKa = 10.05AANSMWPDD43 pKa = 3.88LQPGWGFTSPCPPCPVASCQFLLAQSSLDD72 pKa = 3.7VEE74 pKa = 4.69QTSAPPTYY82 pKa = 10.08KK83 pKa = 10.82GLILDD88 pKa = 4.3LWGLASSDD96 pKa = 3.18AKK98 pKa = 10.37TSFSAACLSLQNSWASTAVFLSRR121 pKa = 11.84GFEE124 pKa = 3.78TAFRR128 pKa = 11.84HH129 pKa = 5.38LCWLAIYY136 pKa = 9.7VWYY139 pKa = 10.67SLLRR143 pKa = 11.84SIIVTSARR151 pKa = 11.84FILNNSILALAGTSLFVCTALLVKK175 pKa = 9.99LVRR178 pKa = 11.84WIFGLMPIYY187 pKa = 10.6LVFTFPISVGRR198 pKa = 11.84WICQKK203 pKa = 10.44RR204 pKa = 11.84FWGKK208 pKa = 9.38FSRR211 pKa = 11.84NYY213 pKa = 10.79AEE215 pKa = 5.03EE216 pKa = 4.29KK217 pKa = 10.24CCEE220 pKa = 3.95QFLSFTINQDD230 pKa = 3.45PPKK233 pKa = 10.09KK234 pKa = 9.83CQLVFQKK241 pKa = 10.82EE242 pKa = 4.32DD243 pKa = 3.48GKK245 pKa = 10.22HH246 pKa = 5.82AGYY249 pKa = 7.72GTCVRR254 pKa = 11.84LFNGTNGLLTAYY266 pKa = 9.62HH267 pKa = 5.92VASNSSKK274 pKa = 10.36VVSTRR279 pKa = 11.84TGNKK283 pKa = 9.29IPLSQFKK290 pKa = 9.91PLIVSPLYY298 pKa = 10.84DD299 pKa = 3.24QVLYY303 pKa = 10.84AGPTEE308 pKa = 4.19WEE310 pKa = 4.19SLLGCKK316 pKa = 9.49GVNFAPAKK324 pKa = 9.46TLGASKK330 pKa = 10.75CNIYY334 pKa = 10.22HH335 pKa = 5.6IQKK338 pKa = 9.42NGKK341 pKa = 7.99WGCTNAEE348 pKa = 3.74IEE350 pKa = 4.46GQLKK354 pKa = 9.79HH355 pKa = 6.95CEE357 pKa = 3.85GNRR360 pKa = 11.84XXKK363 pKa = 9.93HH364 pKa = 6.74CEE366 pKa = 3.58GNRR369 pKa = 11.84DD370 pKa = 3.71VLPGQLSVLSNTEE383 pKa = 4.01PGQSGAGYY391 pKa = 10.32FNGKK395 pKa = 6.52TLVAIHH401 pKa = 6.09VGGSLEE407 pKa = 5.14RR408 pKa = 11.84EE409 pKa = 4.34DD410 pKa = 4.69SCATYY415 pKa = 10.84NVAVPVLPKK424 pKa = 10.38PGLTSPHH431 pKa = 5.96YY432 pKa = 10.56VFEE435 pKa = 4.55TTAPTSGVYY444 pKa = 9.8DD445 pKa = 3.6SKK447 pKa = 11.34IFTALDD453 pKa = 3.49EE454 pKa = 4.57AVEE457 pKa = 4.01QAEE460 pKa = 4.2RR461 pKa = 11.84WVKK464 pKa = 9.62MKK466 pKa = 10.67LSKK469 pKa = 11.43GEE471 pKa = 4.17TLWADD476 pKa = 3.95LEE478 pKa = 4.93DD479 pKa = 4.42DD480 pKa = 4.15LPYY483 pKa = 10.7EE484 pKa = 4.25VEE486 pKa = 4.18TNKK489 pKa = 8.97TAKK492 pKa = 10.23APPSSSSPTEE502 pKa = 3.61TSLDD506 pKa = 3.76CEE508 pKa = 4.23QSGKK512 pKa = 10.03RR513 pKa = 11.84DD514 pKa = 3.08ARR516 pKa = 11.84LRR518 pKa = 11.84PRR520 pKa = 11.84NNRR523 pKa = 11.84KK524 pKa = 9.66DD525 pKa = 3.38RR526 pKa = 11.84IKK528 pKa = 10.22PRR530 pKa = 11.84KK531 pKa = 9.02RR532 pKa = 11.84GLRR535 pKa = 11.84HH536 pKa = 5.17PSKK539 pKa = 10.65RR540 pKa = 11.84YY541 pKa = 10.14ADD543 pKa = 3.06WGTNNEE549 pKa = 4.35DD550 pKa = 3.09VGGEE554 pKa = 3.99NRR556 pKa = 11.84FQYY559 pKa = 10.09QPEE562 pKa = 3.87RR563 pKa = 11.84SRR565 pKa = 11.84EE566 pKa = 4.17GFCGSDD572 pKa = 2.79QGTGPEE578 pKa = 4.01EE579 pKa = 4.19TKK581 pKa = 10.76PVITPAAKK589 pKa = 9.71QEE591 pKa = 4.33RR592 pKa = 11.84RR593 pKa = 11.84QQWIAKK599 pKa = 6.23TNNYY603 pKa = 9.47RR604 pKa = 11.84RR605 pKa = 11.84FFDD608 pKa = 3.93SQFNWEE614 pKa = 4.29VCPSEE619 pKa = 4.17QEE621 pKa = 3.92EE622 pKa = 4.42VAGFRR627 pKa = 11.84FCGKK631 pKa = 10.08APQWYY636 pKa = 9.17HH637 pKa = 6.45PKK639 pKa = 10.11QKK641 pKa = 10.42QEE643 pKa = 4.15GGWGEE648 pKa = 4.3EE649 pKa = 4.2VCRR652 pKa = 11.84XHH654 pKa = 7.74PEE656 pKa = 3.94MGEE659 pKa = 4.07KK660 pKa = 8.78THH662 pKa = 6.72GFGWPQFGPQAEE674 pKa = 4.44LKK676 pKa = 10.49SLRR679 pKa = 11.84LQAARR684 pKa = 11.84WLQRR688 pKa = 11.84AQSAEE693 pKa = 4.15VPPLPEE699 pKa = 3.77RR700 pKa = 11.84EE701 pKa = 3.84RR702 pKa = 11.84VIKK705 pKa = 10.55RR706 pKa = 11.84LVSAYY711 pKa = 9.64KK712 pKa = 9.77QAQSINPVATMSGEE726 pKa = 4.18LLWEE730 pKa = 4.65DD731 pKa = 3.57FLEE734 pKa = 4.39SFKK737 pKa = 11.06EE738 pKa = 4.23AVSSLQLDD746 pKa = 3.63AGVGVPYY753 pKa = 10.48VALGKK758 pKa = 7.54PTHH761 pKa = 6.73RR762 pKa = 11.84SLVEE766 pKa = 4.24DD767 pKa = 4.17PEE769 pKa = 4.25MLPVLARR776 pKa = 11.84LTFDD780 pKa = 3.9RR781 pKa = 11.84LEE783 pKa = 4.28KK784 pKa = 10.39LSKK787 pKa = 11.34GEE789 pKa = 3.81ICRR792 pKa = 11.84LTPEE796 pKa = 3.87EE797 pKa = 4.25LVRR800 pKa = 11.84EE801 pKa = 5.51GYY803 pKa = 10.29CDD805 pKa = 4.76PIRR808 pKa = 11.84VFVKK812 pKa = 10.78GEE814 pKa = 3.59PHH816 pKa = 6.19KK817 pKa = 10.54QSKK820 pKa = 10.21LDD822 pKa = 3.38EE823 pKa = 4.27GRR825 pKa = 11.84YY826 pKa = 8.96RR827 pKa = 11.84LIMSVSLVDD836 pKa = 3.45QLVARR841 pKa = 11.84VLFQEE846 pKa = 4.22QNKK849 pKa = 9.84KK850 pKa = 10.25EE851 pKa = 3.95IQLWRR856 pKa = 11.84VVPSKK861 pKa = 10.72PGFGLSTDD869 pKa = 3.72GQVLEE874 pKa = 4.38FTEE877 pKa = 4.52ALAHH881 pKa = 6.1KK882 pKa = 10.54VGVTPQDD889 pKa = 4.47LIEE892 pKa = 4.2NWKK895 pKa = 10.36QYY897 pKa = 10.81LVPTDD902 pKa = 3.64CSGFDD907 pKa = 3.29WSVADD912 pKa = 4.67WMLDD916 pKa = 3.24DD917 pKa = 6.35DD918 pKa = 4.21IEE920 pKa = 4.34VRR922 pKa = 11.84NRR924 pKa = 11.84LTRR927 pKa = 11.84GLTPVTALLRR937 pKa = 11.84RR938 pKa = 11.84NWKK941 pKa = 8.96HH942 pKa = 5.45CXANSVLCLSDD953 pKa = 3.3GTLLAQCVPGVQKK966 pKa = 10.73SGSYY970 pKa = 8.25NTSSSNSRR978 pKa = 11.84IRR980 pKa = 11.84VMSAYY985 pKa = 10.18HH986 pKa = 7.12CGATWCCAMGDD997 pKa = 3.94DD998 pKa = 4.78ALEE1001 pKa = 4.44SVDD1004 pKa = 5.31SNLEE1008 pKa = 3.79EE1009 pKa = 4.26YY1010 pKa = 10.37KK1011 pKa = 10.82RR1012 pKa = 11.84LGLKK1016 pKa = 10.26VEE1018 pKa = 4.21VSGKK1022 pKa = 10.76LEE1024 pKa = 4.05FCSHH1028 pKa = 6.17IFEE1031 pKa = 4.85SPSLATPVNVGKK1043 pKa = 9.09MLYY1046 pKa = 10.29KK1047 pKa = 10.69LIYY1050 pKa = 9.53GYY1052 pKa = 11.16NPGCEE1057 pKa = 4.03NLEE1060 pKa = 3.91VTANYY1065 pKa = 9.75LAACFSVFNEE1075 pKa = 3.97LRR1077 pKa = 11.84HH1078 pKa = 6.13DD1079 pKa = 4.07PEE1081 pKa = 4.41LVRR1084 pKa = 11.84VLYY1087 pKa = 9.74EE1088 pKa = 3.87WLVLPVQSQNNTT1100 pKa = 3.13
MM1 pKa = 7.53NSFSVLFFFFCAFFCVAPVISGPGPVQPEE30 pKa = 3.76WAASKK35 pKa = 10.05AANSMWPDD43 pKa = 3.88LQPGWGFTSPCPPCPVASCQFLLAQSSLDD72 pKa = 3.7VEE74 pKa = 4.69QTSAPPTYY82 pKa = 10.08KK83 pKa = 10.82GLILDD88 pKa = 4.3LWGLASSDD96 pKa = 3.18AKK98 pKa = 10.37TSFSAACLSLQNSWASTAVFLSRR121 pKa = 11.84GFEE124 pKa = 3.78TAFRR128 pKa = 11.84HH129 pKa = 5.38LCWLAIYY136 pKa = 9.7VWYY139 pKa = 10.67SLLRR143 pKa = 11.84SIIVTSARR151 pKa = 11.84FILNNSILALAGTSLFVCTALLVKK175 pKa = 9.99LVRR178 pKa = 11.84WIFGLMPIYY187 pKa = 10.6LVFTFPISVGRR198 pKa = 11.84WICQKK203 pKa = 10.44RR204 pKa = 11.84FWGKK208 pKa = 9.38FSRR211 pKa = 11.84NYY213 pKa = 10.79AEE215 pKa = 5.03EE216 pKa = 4.29KK217 pKa = 10.24CCEE220 pKa = 3.95QFLSFTINQDD230 pKa = 3.45PPKK233 pKa = 10.09KK234 pKa = 9.83CQLVFQKK241 pKa = 10.82EE242 pKa = 4.32DD243 pKa = 3.48GKK245 pKa = 10.22HH246 pKa = 5.82AGYY249 pKa = 7.72GTCVRR254 pKa = 11.84LFNGTNGLLTAYY266 pKa = 9.62HH267 pKa = 5.92VASNSSKK274 pKa = 10.36VVSTRR279 pKa = 11.84TGNKK283 pKa = 9.29IPLSQFKK290 pKa = 9.91PLIVSPLYY298 pKa = 10.84DD299 pKa = 3.24QVLYY303 pKa = 10.84AGPTEE308 pKa = 4.19WEE310 pKa = 4.19SLLGCKK316 pKa = 9.49GVNFAPAKK324 pKa = 9.46TLGASKK330 pKa = 10.75CNIYY334 pKa = 10.22HH335 pKa = 5.6IQKK338 pKa = 9.42NGKK341 pKa = 7.99WGCTNAEE348 pKa = 3.74IEE350 pKa = 4.46GQLKK354 pKa = 9.79HH355 pKa = 6.95CEE357 pKa = 3.85GNRR360 pKa = 11.84XXKK363 pKa = 9.93HH364 pKa = 6.74CEE366 pKa = 3.58GNRR369 pKa = 11.84DD370 pKa = 3.71VLPGQLSVLSNTEE383 pKa = 4.01PGQSGAGYY391 pKa = 10.32FNGKK395 pKa = 6.52TLVAIHH401 pKa = 6.09VGGSLEE407 pKa = 5.14RR408 pKa = 11.84EE409 pKa = 4.34DD410 pKa = 4.69SCATYY415 pKa = 10.84NVAVPVLPKK424 pKa = 10.38PGLTSPHH431 pKa = 5.96YY432 pKa = 10.56VFEE435 pKa = 4.55TTAPTSGVYY444 pKa = 9.8DD445 pKa = 3.6SKK447 pKa = 11.34IFTALDD453 pKa = 3.49EE454 pKa = 4.57AVEE457 pKa = 4.01QAEE460 pKa = 4.2RR461 pKa = 11.84WVKK464 pKa = 9.62MKK466 pKa = 10.67LSKK469 pKa = 11.43GEE471 pKa = 4.17TLWADD476 pKa = 3.95LEE478 pKa = 4.93DD479 pKa = 4.42DD480 pKa = 4.15LPYY483 pKa = 10.7EE484 pKa = 4.25VEE486 pKa = 4.18TNKK489 pKa = 8.97TAKK492 pKa = 10.23APPSSSSPTEE502 pKa = 3.61TSLDD506 pKa = 3.76CEE508 pKa = 4.23QSGKK512 pKa = 10.03RR513 pKa = 11.84DD514 pKa = 3.08ARR516 pKa = 11.84LRR518 pKa = 11.84PRR520 pKa = 11.84NNRR523 pKa = 11.84KK524 pKa = 9.66DD525 pKa = 3.38RR526 pKa = 11.84IKK528 pKa = 10.22PRR530 pKa = 11.84KK531 pKa = 9.02RR532 pKa = 11.84GLRR535 pKa = 11.84HH536 pKa = 5.17PSKK539 pKa = 10.65RR540 pKa = 11.84YY541 pKa = 10.14ADD543 pKa = 3.06WGTNNEE549 pKa = 4.35DD550 pKa = 3.09VGGEE554 pKa = 3.99NRR556 pKa = 11.84FQYY559 pKa = 10.09QPEE562 pKa = 3.87RR563 pKa = 11.84SRR565 pKa = 11.84EE566 pKa = 4.17GFCGSDD572 pKa = 2.79QGTGPEE578 pKa = 4.01EE579 pKa = 4.19TKK581 pKa = 10.76PVITPAAKK589 pKa = 9.71QEE591 pKa = 4.33RR592 pKa = 11.84RR593 pKa = 11.84QQWIAKK599 pKa = 6.23TNNYY603 pKa = 9.47RR604 pKa = 11.84RR605 pKa = 11.84FFDD608 pKa = 3.93SQFNWEE614 pKa = 4.29VCPSEE619 pKa = 4.17QEE621 pKa = 3.92EE622 pKa = 4.42VAGFRR627 pKa = 11.84FCGKK631 pKa = 10.08APQWYY636 pKa = 9.17HH637 pKa = 6.45PKK639 pKa = 10.11QKK641 pKa = 10.42QEE643 pKa = 4.15GGWGEE648 pKa = 4.3EE649 pKa = 4.2VCRR652 pKa = 11.84XHH654 pKa = 7.74PEE656 pKa = 3.94MGEE659 pKa = 4.07KK660 pKa = 8.78THH662 pKa = 6.72GFGWPQFGPQAEE674 pKa = 4.44LKK676 pKa = 10.49SLRR679 pKa = 11.84LQAARR684 pKa = 11.84WLQRR688 pKa = 11.84AQSAEE693 pKa = 4.15VPPLPEE699 pKa = 3.77RR700 pKa = 11.84EE701 pKa = 3.84RR702 pKa = 11.84VIKK705 pKa = 10.55RR706 pKa = 11.84LVSAYY711 pKa = 9.64KK712 pKa = 9.77QAQSINPVATMSGEE726 pKa = 4.18LLWEE730 pKa = 4.65DD731 pKa = 3.57FLEE734 pKa = 4.39SFKK737 pKa = 11.06EE738 pKa = 4.23AVSSLQLDD746 pKa = 3.63AGVGVPYY753 pKa = 10.48VALGKK758 pKa = 7.54PTHH761 pKa = 6.73RR762 pKa = 11.84SLVEE766 pKa = 4.24DD767 pKa = 4.17PEE769 pKa = 4.25MLPVLARR776 pKa = 11.84LTFDD780 pKa = 3.9RR781 pKa = 11.84LEE783 pKa = 4.28KK784 pKa = 10.39LSKK787 pKa = 11.34GEE789 pKa = 3.81ICRR792 pKa = 11.84LTPEE796 pKa = 3.87EE797 pKa = 4.25LVRR800 pKa = 11.84EE801 pKa = 5.51GYY803 pKa = 10.29CDD805 pKa = 4.76PIRR808 pKa = 11.84VFVKK812 pKa = 10.78GEE814 pKa = 3.59PHH816 pKa = 6.19KK817 pKa = 10.54QSKK820 pKa = 10.21LDD822 pKa = 3.38EE823 pKa = 4.27GRR825 pKa = 11.84YY826 pKa = 8.96RR827 pKa = 11.84LIMSVSLVDD836 pKa = 3.45QLVARR841 pKa = 11.84VLFQEE846 pKa = 4.22QNKK849 pKa = 9.84KK850 pKa = 10.25EE851 pKa = 3.95IQLWRR856 pKa = 11.84VVPSKK861 pKa = 10.72PGFGLSTDD869 pKa = 3.72GQVLEE874 pKa = 4.38FTEE877 pKa = 4.52ALAHH881 pKa = 6.1KK882 pKa = 10.54VGVTPQDD889 pKa = 4.47LIEE892 pKa = 4.2NWKK895 pKa = 10.36QYY897 pKa = 10.81LVPTDD902 pKa = 3.64CSGFDD907 pKa = 3.29WSVADD912 pKa = 4.67WMLDD916 pKa = 3.24DD917 pKa = 6.35DD918 pKa = 4.21IEE920 pKa = 4.34VRR922 pKa = 11.84NRR924 pKa = 11.84LTRR927 pKa = 11.84GLTPVTALLRR937 pKa = 11.84RR938 pKa = 11.84NWKK941 pKa = 8.96HH942 pKa = 5.45CXANSVLCLSDD953 pKa = 3.3GTLLAQCVPGVQKK966 pKa = 10.73SGSYY970 pKa = 8.25NTSSSNSRR978 pKa = 11.84IRR980 pKa = 11.84VMSAYY985 pKa = 10.18HH986 pKa = 7.12CGATWCCAMGDD997 pKa = 3.94DD998 pKa = 4.78ALEE1001 pKa = 4.44SVDD1004 pKa = 5.31SNLEE1008 pKa = 3.79EE1009 pKa = 4.26YY1010 pKa = 10.37KK1011 pKa = 10.82RR1012 pKa = 11.84LGLKK1016 pKa = 10.26VEE1018 pKa = 4.21VSGKK1022 pKa = 10.76LEE1024 pKa = 4.05FCSHH1028 pKa = 6.17IFEE1031 pKa = 4.85SPSLATPVNVGKK1043 pKa = 9.09MLYY1046 pKa = 10.29KK1047 pKa = 10.69LIYY1050 pKa = 9.53GYY1052 pKa = 11.16NPGCEE1057 pKa = 4.03NLEE1060 pKa = 3.91VTANYY1065 pKa = 9.75LAACFSVFNEE1075 pKa = 3.97LRR1077 pKa = 11.84HH1078 pKa = 6.13DD1079 pKa = 4.07PEE1081 pKa = 4.41LVRR1084 pKa = 11.84VLYY1087 pKa = 9.74EE1088 pKa = 3.87WLVLPVQSQNNTT1100 pKa = 3.13
Molecular weight: 122.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7T1C3P2|A0A7T1C3P2_9LUTE P3-P5 fusion protein OS=Phasey bean mild yellows virus OX=1756832 GN=RTD PE=4 SV=1
MM1 pKa = 6.44NTVVARR7 pKa = 11.84NNGMGRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.59RR16 pKa = 11.84RR17 pKa = 11.84NRR19 pKa = 11.84RR20 pKa = 11.84VTRR23 pKa = 11.84RR24 pKa = 11.84QRR26 pKa = 11.84VVVVQAPGLPRR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84RR41 pKa = 11.84QRR43 pKa = 11.84RR44 pKa = 11.84NRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84ASRR51 pKa = 11.84GGRR54 pKa = 11.84AGGGRR59 pKa = 11.84SSEE62 pKa = 4.23TFVLSKK68 pKa = 11.29DD69 pKa = 3.47NLAGSSSGSITFGPSLSEE87 pKa = 3.84KK88 pKa = 10.46PEE90 pKa = 3.95FSSGILKK97 pKa = 10.28AYY99 pKa = 10.25HH100 pKa = 6.63EE101 pKa = 4.49YY102 pKa = 10.72KK103 pKa = 10.29IXXXXLEE110 pKa = 5.38FISEE114 pKa = 4.26ASXTSSGSIAYY125 pKa = 9.43EE126 pKa = 3.79LDD128 pKa = 3.31PHH130 pKa = 6.88CKK132 pKa = 9.85YY133 pKa = 11.04SSLQSSINKK142 pKa = 9.84FGITKK147 pKa = 10.16NGSRR151 pKa = 11.84SWSSKK156 pKa = 9.9FINGEE161 pKa = 3.83EE162 pKa = 3.87WHH164 pKa = 7.19DD165 pKa = 3.55ASEE168 pKa = 4.12DD169 pKa = 3.52QFRR172 pKa = 11.84ILYY175 pKa = 8.98KK176 pKa = 11.06GNGASSIAGSFRR188 pKa = 11.84ITFKK192 pKa = 11.13CQFQNPKK199 pKa = 10.32
MM1 pKa = 6.44NTVVARR7 pKa = 11.84NNGMGRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.59RR16 pKa = 11.84RR17 pKa = 11.84NRR19 pKa = 11.84RR20 pKa = 11.84VTRR23 pKa = 11.84RR24 pKa = 11.84QRR26 pKa = 11.84VVVVQAPGLPRR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84RR41 pKa = 11.84QRR43 pKa = 11.84RR44 pKa = 11.84NRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84ASRR51 pKa = 11.84GGRR54 pKa = 11.84AGGGRR59 pKa = 11.84SSEE62 pKa = 4.23TFVLSKK68 pKa = 11.29DD69 pKa = 3.47NLAGSSSGSITFGPSLSEE87 pKa = 3.84KK88 pKa = 10.46PEE90 pKa = 3.95FSSGILKK97 pKa = 10.28AYY99 pKa = 10.25HH100 pKa = 6.63EE101 pKa = 4.49YY102 pKa = 10.72KK103 pKa = 10.29IXXXXLEE110 pKa = 5.38FISEE114 pKa = 4.26ASXTSSGSIAYY125 pKa = 9.43EE126 pKa = 3.79LDD128 pKa = 3.31PHH130 pKa = 6.88CKK132 pKa = 9.85YY133 pKa = 11.04SSLQSSINKK142 pKa = 9.84FGITKK147 pKa = 10.16NGSRR151 pKa = 11.84SWSSKK156 pKa = 9.9FINGEE161 pKa = 3.83EE162 pKa = 3.87WHH164 pKa = 7.19DD165 pKa = 3.55ASEE168 pKa = 4.12DD169 pKa = 3.52QFRR172 pKa = 11.84ILYY175 pKa = 8.98KK176 pKa = 11.06GNGASSIAGSFRR188 pKa = 11.84ITFKK192 pKa = 11.13CQFQNPKK199 pKa = 10.32
Molecular weight: 21.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3144 |
45 |
1100 |
449.1 |
49.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.997 ± 0.459 | 2.449 ± 0.497 |
3.658 ± 0.54 | 5.98 ± 0.54 |
4.23 ± 0.474 | 7.729 ± 0.372 |
1.527 ± 0.207 | 3.658 ± 0.392 |
5.503 ± 0.764 | 8.524 ± 1.225 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.24 ± 0.139 | 4.676 ± 0.376 |
6.266 ± 0.523 | 3.658 ± 0.403 |
6.743 ± 1.161 | 9.733 ± 0.79 |
5.057 ± 0.432 | 6.266 ± 0.516 |
2.036 ± 0.372 | 2.99 ± 0.236 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
