
Halomonas urumqiensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
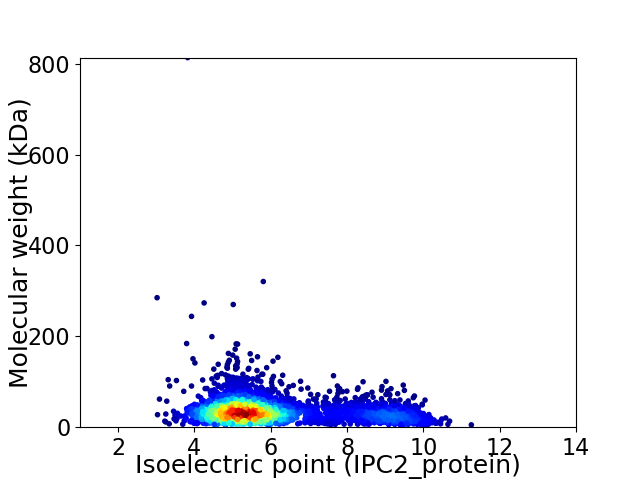
Virtual 2D-PAGE plot for 3488 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N7UI90|A0A2N7UI90_9GAMM 50S ribosomal protein L30 OS=Halomonas urumqiensis OX=1684789 GN=rpmD PE=3 SV=1
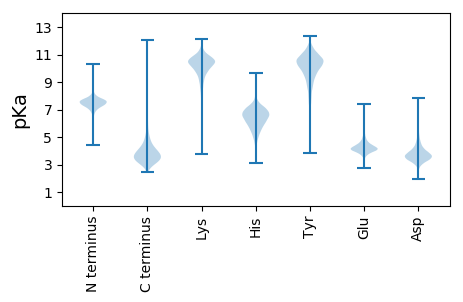
MM1 pKa = 7.17VKK3 pKa = 10.14PMRR6 pKa = 11.84GGGMSSRR13 pKa = 11.84GDD15 pKa = 3.24QRR17 pKa = 11.84GAALVVTLVLLVVALMLGLSSFQSARR43 pKa = 11.84LEE45 pKa = 3.75EE46 pKa = 4.51SMAGNQRR53 pKa = 11.84ASVQALLAAEE63 pKa = 4.23YY64 pKa = 9.82GASKK68 pKa = 10.6AWLEE72 pKa = 4.13FDD74 pKa = 3.87PSSLSSEE81 pKa = 4.15EE82 pKa = 4.16GVLVSDD88 pKa = 5.38DD89 pKa = 4.0RR90 pKa = 11.84DD91 pKa = 3.36NVYY94 pKa = 10.86YY95 pKa = 10.15SWSVTPDD102 pKa = 2.96GYY104 pKa = 11.59GKK106 pKa = 10.69YY107 pKa = 10.18LFNSVGEE114 pKa = 4.23VMAGSDD120 pKa = 4.64LISQRR125 pKa = 11.84VISYY129 pKa = 10.48LGILSIGPGGTIVAANNDD147 pKa = 3.92DD148 pKa = 3.83DD149 pKa = 5.41CEE151 pKa = 4.16EE152 pKa = 4.61GSVEE156 pKa = 4.32FVPPSSNMEE165 pKa = 3.86VTGEE169 pKa = 4.11EE170 pKa = 4.16EE171 pKa = 4.17VNGNIKK177 pKa = 10.49AAVQVGCGIIAEE189 pKa = 4.86NIASSIIPKK198 pKa = 10.31NGEE201 pKa = 3.29FDD203 pKa = 4.17DD204 pKa = 4.85YY205 pKa = 12.0VMHH208 pKa = 7.27DD209 pKa = 4.21TGDD212 pKa = 3.56DD213 pKa = 3.69TYY215 pKa = 11.23TCLDD219 pKa = 3.26GGGNNRR225 pKa = 11.84LCNYY229 pKa = 9.87YY230 pKa = 10.75GGIQGGIDD238 pKa = 4.5LDD240 pKa = 3.67ILKK243 pKa = 10.26NADD246 pKa = 3.11MLAQFVYY253 pKa = 10.32GIRR256 pKa = 11.84DD257 pKa = 3.53NYY259 pKa = 10.73SEE261 pKa = 5.18SIVNSIPSSVPEE273 pKa = 3.99GSITYY278 pKa = 9.68VKK280 pKa = 10.61SADD283 pKa = 3.66DD284 pKa = 3.65ADD286 pKa = 4.23GNRR289 pKa = 11.84EE290 pKa = 3.93TFSRR294 pKa = 11.84SGNFTGSGILIVDD307 pKa = 4.01GNVDD311 pKa = 4.17FGGVPGFEE319 pKa = 3.96GLVIVLGDD327 pKa = 3.65YY328 pKa = 10.18VVSGGGGGEE337 pKa = 4.07FKK339 pKa = 10.91GSVVSAPIVDD349 pKa = 4.13SACWDD354 pKa = 3.26AGAATGCEE362 pKa = 3.91FDD364 pKa = 4.69DD365 pKa = 4.55KK366 pKa = 11.02KK367 pKa = 11.17IEE369 pKa = 4.0IGGGGSSSYY378 pKa = 11.13SYY380 pKa = 11.5DD381 pKa = 3.11PLAILNAFEE390 pKa = 5.76LIDD393 pKa = 4.47GSGLDD398 pKa = 4.15EE399 pKa = 4.64LWGVDD404 pKa = 3.3NQTLKK409 pKa = 11.29GNFYY413 pKa = 8.59MASWSEE419 pKa = 4.18LIGNYY424 pKa = 9.62
MM1 pKa = 7.17VKK3 pKa = 10.14PMRR6 pKa = 11.84GGGMSSRR13 pKa = 11.84GDD15 pKa = 3.24QRR17 pKa = 11.84GAALVVTLVLLVVALMLGLSSFQSARR43 pKa = 11.84LEE45 pKa = 3.75EE46 pKa = 4.51SMAGNQRR53 pKa = 11.84ASVQALLAAEE63 pKa = 4.23YY64 pKa = 9.82GASKK68 pKa = 10.6AWLEE72 pKa = 4.13FDD74 pKa = 3.87PSSLSSEE81 pKa = 4.15EE82 pKa = 4.16GVLVSDD88 pKa = 5.38DD89 pKa = 4.0RR90 pKa = 11.84DD91 pKa = 3.36NVYY94 pKa = 10.86YY95 pKa = 10.15SWSVTPDD102 pKa = 2.96GYY104 pKa = 11.59GKK106 pKa = 10.69YY107 pKa = 10.18LFNSVGEE114 pKa = 4.23VMAGSDD120 pKa = 4.64LISQRR125 pKa = 11.84VISYY129 pKa = 10.48LGILSIGPGGTIVAANNDD147 pKa = 3.92DD148 pKa = 3.83DD149 pKa = 5.41CEE151 pKa = 4.16EE152 pKa = 4.61GSVEE156 pKa = 4.32FVPPSSNMEE165 pKa = 3.86VTGEE169 pKa = 4.11EE170 pKa = 4.16EE171 pKa = 4.17VNGNIKK177 pKa = 10.49AAVQVGCGIIAEE189 pKa = 4.86NIASSIIPKK198 pKa = 10.31NGEE201 pKa = 3.29FDD203 pKa = 4.17DD204 pKa = 4.85YY205 pKa = 12.0VMHH208 pKa = 7.27DD209 pKa = 4.21TGDD212 pKa = 3.56DD213 pKa = 3.69TYY215 pKa = 11.23TCLDD219 pKa = 3.26GGGNNRR225 pKa = 11.84LCNYY229 pKa = 9.87YY230 pKa = 10.75GGIQGGIDD238 pKa = 4.5LDD240 pKa = 3.67ILKK243 pKa = 10.26NADD246 pKa = 3.11MLAQFVYY253 pKa = 10.32GIRR256 pKa = 11.84DD257 pKa = 3.53NYY259 pKa = 10.73SEE261 pKa = 5.18SIVNSIPSSVPEE273 pKa = 3.99GSITYY278 pKa = 9.68VKK280 pKa = 10.61SADD283 pKa = 3.66DD284 pKa = 3.65ADD286 pKa = 4.23GNRR289 pKa = 11.84EE290 pKa = 3.93TFSRR294 pKa = 11.84SGNFTGSGILIVDD307 pKa = 4.01GNVDD311 pKa = 4.17FGGVPGFEE319 pKa = 3.96GLVIVLGDD327 pKa = 3.65YY328 pKa = 10.18VVSGGGGGEE337 pKa = 4.07FKK339 pKa = 10.91GSVVSAPIVDD349 pKa = 4.13SACWDD354 pKa = 3.26AGAATGCEE362 pKa = 3.91FDD364 pKa = 4.69DD365 pKa = 4.55KK366 pKa = 11.02KK367 pKa = 11.17IEE369 pKa = 4.0IGGGGSSSYY378 pKa = 11.13SYY380 pKa = 11.5DD381 pKa = 3.11PLAILNAFEE390 pKa = 5.76LIDD393 pKa = 4.47GSGLDD398 pKa = 4.15EE399 pKa = 4.64LWGVDD404 pKa = 3.3NQTLKK409 pKa = 11.29GNFYY413 pKa = 8.59MASWSEE419 pKa = 4.18LIGNYY424 pKa = 9.62
Molecular weight: 44.47 kDa
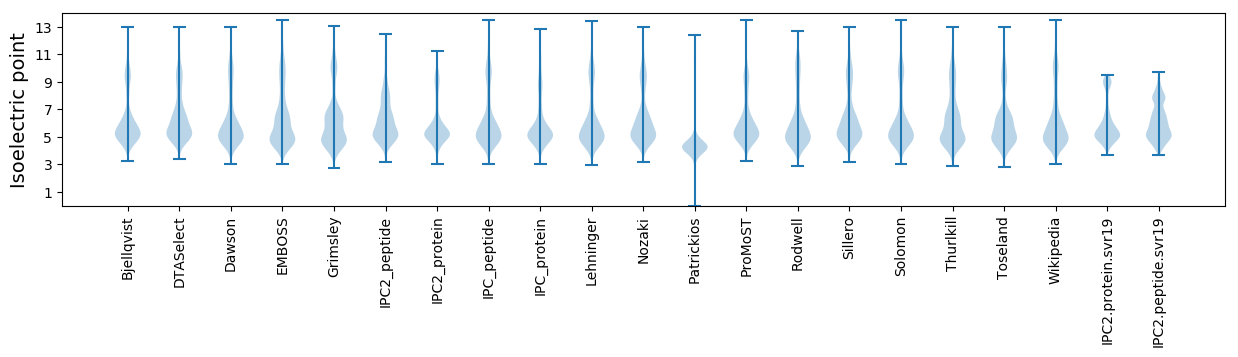
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N7UK81|A0A2N7UK81_9GAMM 1-deoxy-D-xylulose-5-phosphate synthase OS=Halomonas urumqiensis OX=1684789 GN=dxs PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1180462 |
37 |
7991 |
338.4 |
37.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.274 ± 0.053 | 0.903 ± 0.013 |
5.919 ± 0.043 | 6.489 ± 0.044 |
3.424 ± 0.027 | 8.433 ± 0.045 |
2.504 ± 0.025 | 4.707 ± 0.033 |
2.46 ± 0.037 | 11.459 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.569 ± 0.025 | 2.518 ± 0.026 |
4.931 ± 0.033 | 3.737 ± 0.028 |
7.223 ± 0.048 | 5.489 ± 0.03 |
4.993 ± 0.036 | 7.31 ± 0.035 |
1.446 ± 0.018 | 2.213 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
