
Desulfocucumis palustris
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Peptococcaceae; Desulfocucumis
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
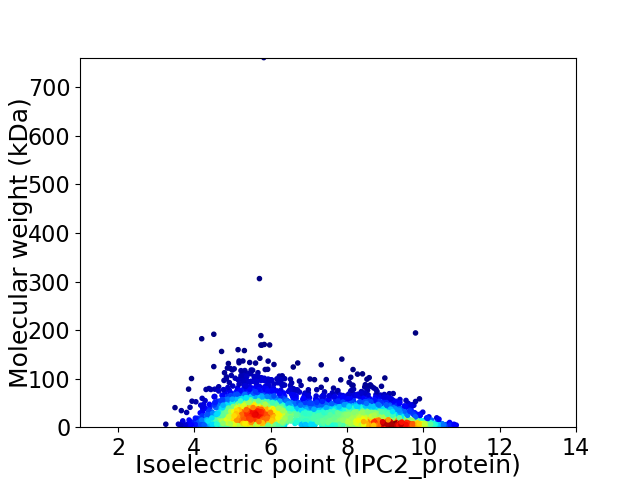
Virtual 2D-PAGE plot for 3959 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L2XM15|A0A2L2XM15_9FIRM Uncharacterized protein OS=Desulfocucumis palustris OX=1898651 GN=DCCM_4112 PE=4 SV=1
MM1 pKa = 7.54AKK3 pKa = 10.07KK4 pKa = 9.73PRR6 pKa = 11.84NPWCIRR12 pKa = 11.84YY13 pKa = 9.95ANIGYY18 pKa = 7.0YY19 pKa = 9.77HH20 pKa = 7.64RR21 pKa = 11.84NTVCLSANPAAVTVEE36 pKa = 3.88ANQTEE41 pKa = 4.59NVALTAEE48 pKa = 4.38KK49 pKa = 10.56DD50 pKa = 3.76GEE52 pKa = 4.48VVEE55 pKa = 4.9GVDD58 pKa = 4.26YY59 pKa = 11.22DD60 pKa = 3.92VTSADD65 pKa = 3.62EE66 pKa = 4.4TVATATEE73 pKa = 4.11ADD75 pKa = 3.74GVITVTGVRR84 pKa = 11.84EE85 pKa = 4.24GSTEE89 pKa = 4.1LTVTAAKK96 pKa = 10.25EE97 pKa = 4.29GEE99 pKa = 4.42EE100 pKa = 4.21FAAIAPIAVTVTEE113 pKa = 4.24GTIATVEE120 pKa = 3.91ITGAASVPVGTSVDD134 pKa = 3.63LTAVAKK140 pKa = 10.49DD141 pKa = 3.09AGGNVIADD149 pKa = 4.13AGAVTWAITSDD160 pKa = 3.51NADD163 pKa = 3.48NAVLTQSGKK172 pKa = 10.71FIATVAGSYY181 pKa = 9.06TVTATIGAVVGTATIQVSGIAAGVTFTASKK211 pKa = 9.49TSVVANATDD220 pKa = 3.41TADD223 pKa = 3.17ITVSVVDD230 pKa = 3.63TNGNVVTNFNGTAAPQRR247 pKa = 11.84TPVAGAQAVAVPATVTITNGVGTFTATSFAAGSIGTDD284 pKa = 3.08SLSITVGTLVSTNGQPIATSLNYY307 pKa = 10.89GNVQVSSVARR317 pKa = 11.84TATALTLSGPAYY329 pKa = 10.53VSANGNTQATLNVGYY344 pKa = 10.75ADD346 pKa = 3.76QNGAAMNAPAAQYY359 pKa = 8.23VTYY362 pKa = 9.63TLTGPASFQQGGAPVTTQSVFVPAAGGTVNIFAIMGQTGAITLTASAAGLTTATAQIQSVINTAATDD429 pKa = 3.36ITVTSEE435 pKa = 4.0TATLTAVCNADD446 pKa = 3.27NAGTNLPAGTAFNRR460 pKa = 11.84ITIQLVDD467 pKa = 3.77TNGLPVAGTDD477 pKa = 4.08ALNISDD483 pKa = 4.02NTTAASSLYY492 pKa = 9.61YY493 pKa = 8.63WTVNANGQPGVYY505 pKa = 10.01LGTTPQVSALVGGRR519 pKa = 11.84LQLAVLNTAVAATSPTITVRR539 pKa = 11.84DD540 pKa = 3.63TVLNVAKK547 pKa = 7.48TTTYY551 pKa = 11.0SYY553 pKa = 9.3VTSSASTATWVGQAIQTAMAGQSVTLTLQLQDD585 pKa = 3.0IAGNDD590 pKa = 3.45LQIANRR596 pKa = 11.84NVPVYY601 pKa = 10.08FLANAANATIAGSSAWTVLNPYY623 pKa = 10.31NVTTNAQGQATVTVAVPAASAGSAFTLTSALTGGVPAVVTVTAVNASAYY672 pKa = 8.59ATGLVLANTAVAAGSIPNPTTEE694 pKa = 4.5VITWPNGNMTAGQNLAAFVGSAINVLPVNAIGQVANATDD733 pKa = 4.36VIRR736 pKa = 11.84VTSSNEE742 pKa = 3.58NVLDD746 pKa = 3.62IAGVTNGFVEE756 pKa = 5.1GPFAVGPVPAITAVNSGTATLTFTNVTNPAVPAVTKK792 pKa = 10.14TITVVPATAPAAVLGMNPDD811 pKa = 3.16GTTNRR816 pKa = 11.84VYY818 pKa = 11.15NFTTAGVVGPFTLQAVDD835 pKa = 3.4GGNNLVAATVPITLTAVQVDD855 pKa = 4.78NILGTPGTPGIRR867 pKa = 11.84TSAAGSNVTSVTIPQGQASVQIWVDD892 pKa = 4.56GITLPSATAATAAVALNTLAPEE914 pKa = 4.21IVSAAINGDD923 pKa = 3.85TLVVTMNNPVTGTATAAAFDD943 pKa = 4.15VVIDD947 pKa = 4.17AAPAVNPTGDD957 pKa = 3.63GVISGNTITLTLAAPVTNANVVTLAYY983 pKa = 6.91TTDD986 pKa = 3.19SFLTTGYY993 pKa = 9.67QAPLATTIAPIAVTNNTPP1011 pKa = 3.02
MM1 pKa = 7.54AKK3 pKa = 10.07KK4 pKa = 9.73PRR6 pKa = 11.84NPWCIRR12 pKa = 11.84YY13 pKa = 9.95ANIGYY18 pKa = 7.0YY19 pKa = 9.77HH20 pKa = 7.64RR21 pKa = 11.84NTVCLSANPAAVTVEE36 pKa = 3.88ANQTEE41 pKa = 4.59NVALTAEE48 pKa = 4.38KK49 pKa = 10.56DD50 pKa = 3.76GEE52 pKa = 4.48VVEE55 pKa = 4.9GVDD58 pKa = 4.26YY59 pKa = 11.22DD60 pKa = 3.92VTSADD65 pKa = 3.62EE66 pKa = 4.4TVATATEE73 pKa = 4.11ADD75 pKa = 3.74GVITVTGVRR84 pKa = 11.84EE85 pKa = 4.24GSTEE89 pKa = 4.1LTVTAAKK96 pKa = 10.25EE97 pKa = 4.29GEE99 pKa = 4.42EE100 pKa = 4.21FAAIAPIAVTVTEE113 pKa = 4.24GTIATVEE120 pKa = 3.91ITGAASVPVGTSVDD134 pKa = 3.63LTAVAKK140 pKa = 10.49DD141 pKa = 3.09AGGNVIADD149 pKa = 4.13AGAVTWAITSDD160 pKa = 3.51NADD163 pKa = 3.48NAVLTQSGKK172 pKa = 10.71FIATVAGSYY181 pKa = 9.06TVTATIGAVVGTATIQVSGIAAGVTFTASKK211 pKa = 9.49TSVVANATDD220 pKa = 3.41TADD223 pKa = 3.17ITVSVVDD230 pKa = 3.63TNGNVVTNFNGTAAPQRR247 pKa = 11.84TPVAGAQAVAVPATVTITNGVGTFTATSFAAGSIGTDD284 pKa = 3.08SLSITVGTLVSTNGQPIATSLNYY307 pKa = 10.89GNVQVSSVARR317 pKa = 11.84TATALTLSGPAYY329 pKa = 10.53VSANGNTQATLNVGYY344 pKa = 10.75ADD346 pKa = 3.76QNGAAMNAPAAQYY359 pKa = 8.23VTYY362 pKa = 9.63TLTGPASFQQGGAPVTTQSVFVPAAGGTVNIFAIMGQTGAITLTASAAGLTTATAQIQSVINTAATDD429 pKa = 3.36ITVTSEE435 pKa = 4.0TATLTAVCNADD446 pKa = 3.27NAGTNLPAGTAFNRR460 pKa = 11.84ITIQLVDD467 pKa = 3.77TNGLPVAGTDD477 pKa = 4.08ALNISDD483 pKa = 4.02NTTAASSLYY492 pKa = 9.61YY493 pKa = 8.63WTVNANGQPGVYY505 pKa = 10.01LGTTPQVSALVGGRR519 pKa = 11.84LQLAVLNTAVAATSPTITVRR539 pKa = 11.84DD540 pKa = 3.63TVLNVAKK547 pKa = 7.48TTTYY551 pKa = 11.0SYY553 pKa = 9.3VTSSASTATWVGQAIQTAMAGQSVTLTLQLQDD585 pKa = 3.0IAGNDD590 pKa = 3.45LQIANRR596 pKa = 11.84NVPVYY601 pKa = 10.08FLANAANATIAGSSAWTVLNPYY623 pKa = 10.31NVTTNAQGQATVTVAVPAASAGSAFTLTSALTGGVPAVVTVTAVNASAYY672 pKa = 8.59ATGLVLANTAVAAGSIPNPTTEE694 pKa = 4.5VITWPNGNMTAGQNLAAFVGSAINVLPVNAIGQVANATDD733 pKa = 4.36VIRR736 pKa = 11.84VTSSNEE742 pKa = 3.58NVLDD746 pKa = 3.62IAGVTNGFVEE756 pKa = 5.1GPFAVGPVPAITAVNSGTATLTFTNVTNPAVPAVTKK792 pKa = 10.14TITVVPATAPAAVLGMNPDD811 pKa = 3.16GTTNRR816 pKa = 11.84VYY818 pKa = 11.15NFTTAGVVGPFTLQAVDD835 pKa = 3.4GGNNLVAATVPITLTAVQVDD855 pKa = 4.78NILGTPGTPGIRR867 pKa = 11.84TSAAGSNVTSVTIPQGQASVQIWVDD892 pKa = 4.56GITLPSATAATAAVALNTLAPEE914 pKa = 4.21IVSAAINGDD923 pKa = 3.85TLVVTMNNPVTGTATAAAFDD943 pKa = 4.15VVIDD947 pKa = 4.17AAPAVNPTGDD957 pKa = 3.63GVISGNTITLTLAAPVTNANVVTLAYY983 pKa = 6.91TTDD986 pKa = 3.19SFLTTGYY993 pKa = 9.67QAPLATTIAPIAVTNNTPP1011 pKa = 3.02
Molecular weight: 100.24 kDa
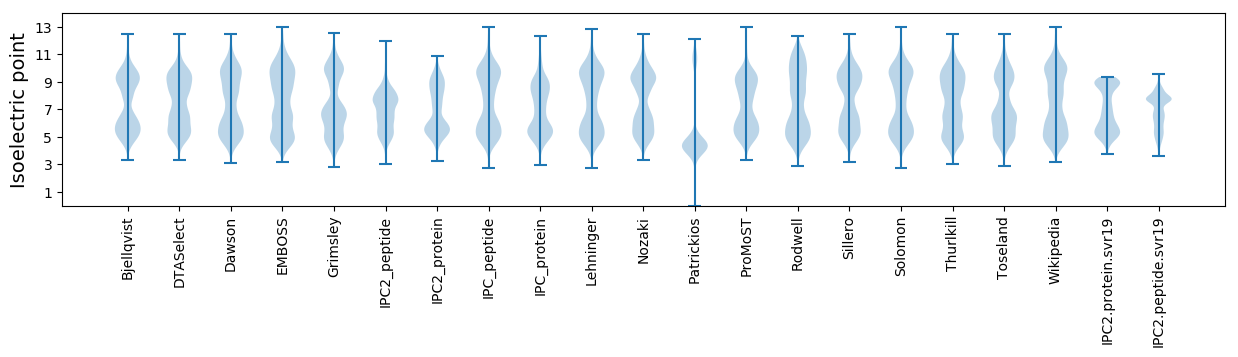
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L2XFA6|A0A2L2XFA6_9FIRM N-acetyltransferase domain-containing protein OS=Desulfocucumis palustris OX=1898651 GN=DCCM_3932 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPKK8 pKa = 8.41NRR10 pKa = 11.84KK11 pKa = 7.86HH12 pKa = 6.2KK13 pKa = 10.07KK14 pKa = 7.69VHH16 pKa = 5.74GFLKK20 pKa = 10.48RR21 pKa = 11.84MSSPGGEE28 pKa = 3.76NVIKK32 pKa = 10.52RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.22GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPKK8 pKa = 8.41NRR10 pKa = 11.84KK11 pKa = 7.86HH12 pKa = 6.2KK13 pKa = 10.07KK14 pKa = 7.69VHH16 pKa = 5.74GFLKK20 pKa = 10.48RR21 pKa = 11.84MSSPGGEE28 pKa = 3.76NVIKK32 pKa = 10.52RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.22GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
Molecular weight: 5.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1035266 |
37 |
6798 |
261.5 |
28.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.264 ± 0.053 | 1.218 ± 0.016 |
4.943 ± 0.031 | 6.741 ± 0.047 |
3.981 ± 0.026 | 8.203 ± 0.045 |
1.674 ± 0.015 | 7.059 ± 0.042 |
5.99 ± 0.038 | 10.03 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.76 ± 0.021 | 4.134 ± 0.032 |
4.304 ± 0.026 | 3.14 ± 0.027 |
5.487 ± 0.043 | 5.68 ± 0.038 |
4.818 ± 0.036 | 7.259 ± 0.036 |
0.996 ± 0.015 | 3.319 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
