
Human papillomavirus 178
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 24
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
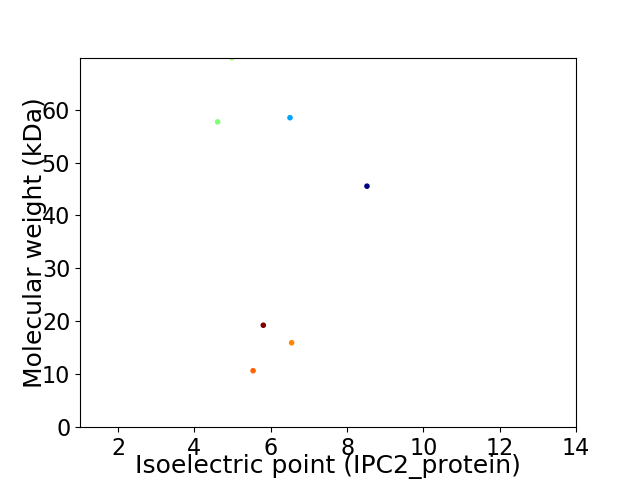
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|X2GCR4|X2GCR4_9PAPI Major capsid protein L1 OS=Human papillomavirus 178 OX=1478160 GN=L1 PE=3 SV=1
MM1 pKa = 7.39TSRR4 pKa = 11.84AKK6 pKa = 10.34RR7 pKa = 11.84IKK9 pKa = 10.02RR10 pKa = 11.84DD11 pKa = 3.55SVEE14 pKa = 4.15NLYY17 pKa = 10.03KK18 pKa = 10.07QCKK21 pKa = 8.31LTGQCPDD28 pKa = 3.27DD29 pKa = 4.54VINKK33 pKa = 9.39VEE35 pKa = 4.16GTTLADD41 pKa = 3.78RR42 pKa = 11.84LLKK45 pKa = 10.5IIGSVVYY52 pKa = 9.95FGGLGIGTGKK62 pKa = 10.77GSGGATGYY70 pKa = 10.55RR71 pKa = 11.84PIGATTPRR79 pKa = 11.84VTDD82 pKa = 5.33AIPVRR87 pKa = 11.84PVVPVDD93 pKa = 3.39PLVPAEE99 pKa = 5.37LIPVDD104 pKa = 4.51PGASSIIPLTEE115 pKa = 4.23AGIPDD120 pKa = 3.45ATLIEE125 pKa = 4.52SGGAVIDD132 pKa = 4.36AGSAEE137 pKa = 4.47PPVLTTIDD145 pKa = 4.64PISDD149 pKa = 3.24VTAVDD154 pKa = 3.59NQPTVITGQDD164 pKa = 3.06EE165 pKa = 4.68NIAVLDD171 pKa = 3.86IQPTPQAPKK180 pKa = 10.07RR181 pKa = 11.84VISGVRR187 pKa = 11.84SLGKK191 pKa = 10.34SPFEE195 pKa = 3.96PQILLPPPDD204 pKa = 3.82MPAIDD209 pKa = 3.36VSVFVDD215 pKa = 3.56AQLVGEE221 pKa = 4.39TVGAEE226 pKa = 4.9LIPLDD231 pKa = 5.04DD232 pKa = 4.02INLIEE237 pKa = 4.32EE238 pKa = 4.43FEE240 pKa = 4.11IQEE243 pKa = 4.21PVTRR247 pKa = 11.84TSTPEE252 pKa = 3.58QRR254 pKa = 11.84LGRR257 pKa = 11.84AFQRR261 pKa = 11.84ARR263 pKa = 11.84EE264 pKa = 4.08LYY266 pKa = 9.71NRR268 pKa = 11.84QIRR271 pKa = 11.84QIRR274 pKa = 11.84TRR276 pKa = 11.84NRR278 pKa = 11.84DD279 pKa = 3.06FLGPVSRR286 pKa = 11.84AIQFDD291 pKa = 3.81FEE293 pKa = 4.41NPAFSTDD300 pKa = 2.78VTLQFDD306 pKa = 3.51QDD308 pKa = 4.14VQNVAATPDD317 pKa = 3.75PDD319 pKa = 3.71FADD322 pKa = 4.28VITVHH327 pKa = 6.71RR328 pKa = 11.84PQYY331 pKa = 10.9SEE333 pKa = 3.9TDD335 pKa = 3.2SGRR338 pKa = 11.84VRR340 pKa = 11.84VSRR343 pKa = 11.84LARR346 pKa = 11.84RR347 pKa = 11.84GTIITRR353 pKa = 11.84SGTQIGEE360 pKa = 4.06NVHH363 pKa = 6.93FYY365 pKa = 11.19FDD367 pKa = 4.28LSTIGSDD374 pKa = 3.0AADD377 pKa = 4.61AIEE380 pKa = 5.07LGTIGQHH387 pKa = 5.6SGEE390 pKa = 4.27TAIVDD395 pKa = 3.84AVAEE399 pKa = 4.43STFVDD404 pKa = 3.58IPNSSEE410 pKa = 3.67IQFDD414 pKa = 4.83DD415 pKa = 4.3EE416 pKa = 5.59DD417 pKa = 4.64LLDD420 pKa = 3.87TQVEE424 pKa = 4.32NFEE427 pKa = 4.52GGHH430 pKa = 6.09LVLSGTGRR438 pKa = 11.84RR439 pKa = 11.84GAVYY443 pKa = 10.19SFPTLAPGVAPKK455 pKa = 10.53IFVDD459 pKa = 4.97DD460 pKa = 3.89FAKK463 pKa = 10.81NLFVSYY469 pKa = 9.8PISHH473 pKa = 6.96NKK475 pKa = 8.19TDD477 pKa = 4.07IIGPDD482 pKa = 3.34TLHH485 pKa = 6.87PLEE488 pKa = 5.09PPIILDD494 pKa = 3.99FQSSTYY500 pKa = 10.05YY501 pKa = 10.44LHH503 pKa = 6.98PSLRR507 pKa = 11.84RR508 pKa = 11.84RR509 pKa = 11.84RR510 pKa = 11.84KK511 pKa = 9.19RR512 pKa = 11.84KK513 pKa = 9.81RR514 pKa = 11.84SDD516 pKa = 2.84IYY518 pKa = 11.55DD519 pKa = 3.71FLSDD523 pKa = 3.89GSVDD527 pKa = 3.55TPEE530 pKa = 3.55WW531 pKa = 3.57
MM1 pKa = 7.39TSRR4 pKa = 11.84AKK6 pKa = 10.34RR7 pKa = 11.84IKK9 pKa = 10.02RR10 pKa = 11.84DD11 pKa = 3.55SVEE14 pKa = 4.15NLYY17 pKa = 10.03KK18 pKa = 10.07QCKK21 pKa = 8.31LTGQCPDD28 pKa = 3.27DD29 pKa = 4.54VINKK33 pKa = 9.39VEE35 pKa = 4.16GTTLADD41 pKa = 3.78RR42 pKa = 11.84LLKK45 pKa = 10.5IIGSVVYY52 pKa = 9.95FGGLGIGTGKK62 pKa = 10.77GSGGATGYY70 pKa = 10.55RR71 pKa = 11.84PIGATTPRR79 pKa = 11.84VTDD82 pKa = 5.33AIPVRR87 pKa = 11.84PVVPVDD93 pKa = 3.39PLVPAEE99 pKa = 5.37LIPVDD104 pKa = 4.51PGASSIIPLTEE115 pKa = 4.23AGIPDD120 pKa = 3.45ATLIEE125 pKa = 4.52SGGAVIDD132 pKa = 4.36AGSAEE137 pKa = 4.47PPVLTTIDD145 pKa = 4.64PISDD149 pKa = 3.24VTAVDD154 pKa = 3.59NQPTVITGQDD164 pKa = 3.06EE165 pKa = 4.68NIAVLDD171 pKa = 3.86IQPTPQAPKK180 pKa = 10.07RR181 pKa = 11.84VISGVRR187 pKa = 11.84SLGKK191 pKa = 10.34SPFEE195 pKa = 3.96PQILLPPPDD204 pKa = 3.82MPAIDD209 pKa = 3.36VSVFVDD215 pKa = 3.56AQLVGEE221 pKa = 4.39TVGAEE226 pKa = 4.9LIPLDD231 pKa = 5.04DD232 pKa = 4.02INLIEE237 pKa = 4.32EE238 pKa = 4.43FEE240 pKa = 4.11IQEE243 pKa = 4.21PVTRR247 pKa = 11.84TSTPEE252 pKa = 3.58QRR254 pKa = 11.84LGRR257 pKa = 11.84AFQRR261 pKa = 11.84ARR263 pKa = 11.84EE264 pKa = 4.08LYY266 pKa = 9.71NRR268 pKa = 11.84QIRR271 pKa = 11.84QIRR274 pKa = 11.84TRR276 pKa = 11.84NRR278 pKa = 11.84DD279 pKa = 3.06FLGPVSRR286 pKa = 11.84AIQFDD291 pKa = 3.81FEE293 pKa = 4.41NPAFSTDD300 pKa = 2.78VTLQFDD306 pKa = 3.51QDD308 pKa = 4.14VQNVAATPDD317 pKa = 3.75PDD319 pKa = 3.71FADD322 pKa = 4.28VITVHH327 pKa = 6.71RR328 pKa = 11.84PQYY331 pKa = 10.9SEE333 pKa = 3.9TDD335 pKa = 3.2SGRR338 pKa = 11.84VRR340 pKa = 11.84VSRR343 pKa = 11.84LARR346 pKa = 11.84RR347 pKa = 11.84GTIITRR353 pKa = 11.84SGTQIGEE360 pKa = 4.06NVHH363 pKa = 6.93FYY365 pKa = 11.19FDD367 pKa = 4.28LSTIGSDD374 pKa = 3.0AADD377 pKa = 4.61AIEE380 pKa = 5.07LGTIGQHH387 pKa = 5.6SGEE390 pKa = 4.27TAIVDD395 pKa = 3.84AVAEE399 pKa = 4.43STFVDD404 pKa = 3.58IPNSSEE410 pKa = 3.67IQFDD414 pKa = 4.83DD415 pKa = 4.3EE416 pKa = 5.59DD417 pKa = 4.64LLDD420 pKa = 3.87TQVEE424 pKa = 4.32NFEE427 pKa = 4.52GGHH430 pKa = 6.09LVLSGTGRR438 pKa = 11.84RR439 pKa = 11.84GAVYY443 pKa = 10.19SFPTLAPGVAPKK455 pKa = 10.53IFVDD459 pKa = 4.97DD460 pKa = 3.89FAKK463 pKa = 10.81NLFVSYY469 pKa = 9.8PISHH473 pKa = 6.96NKK475 pKa = 8.19TDD477 pKa = 4.07IIGPDD482 pKa = 3.34TLHH485 pKa = 6.87PLEE488 pKa = 5.09PPIILDD494 pKa = 3.99FQSSTYY500 pKa = 10.05YY501 pKa = 10.44LHH503 pKa = 6.98PSLRR507 pKa = 11.84RR508 pKa = 11.84RR509 pKa = 11.84RR510 pKa = 11.84KK511 pKa = 9.19RR512 pKa = 11.84KK513 pKa = 9.81RR514 pKa = 11.84SDD516 pKa = 2.84IYY518 pKa = 11.55DD519 pKa = 3.71FLSDD523 pKa = 3.89GSVDD527 pKa = 3.55TPEE530 pKa = 3.55WW531 pKa = 3.57
Molecular weight: 57.68 kDa
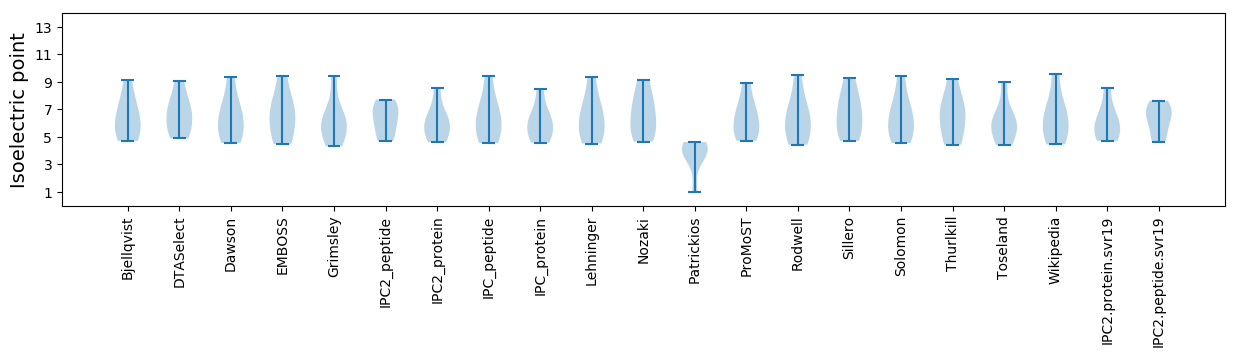
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X2GB93|X2GB93_9PAPI Minor capsid protein L2 OS=Human papillomavirus 178 OX=1478160 GN=L2 PE=3 SV=1
MM1 pKa = 8.0DD2 pKa = 4.22WLTDD6 pKa = 3.53RR7 pKa = 11.84FNAIQDD13 pKa = 3.9NILNLIEE20 pKa = 4.79QGAEE24 pKa = 4.02DD25 pKa = 5.43LDD27 pKa = 4.07SQINYY32 pKa = 7.96WNLVRR37 pKa = 11.84QEE39 pKa = 4.11NVYY42 pKa = 10.55LYY44 pKa = 10.07YY45 pKa = 10.7GRR47 pKa = 11.84KK48 pKa = 9.39EE49 pKa = 4.09GLTHH53 pKa = 7.6FGLQPLPVAAVSEE66 pKa = 4.6YY67 pKa = 10.2KK68 pKa = 10.7AKK70 pKa = 10.48EE71 pKa = 4.24AINMVLLLQSLKK83 pKa = 10.58KK84 pKa = 9.89SAYY87 pKa = 9.85ANEE90 pKa = 3.82VWTLQDD96 pKa = 4.19ASAEE100 pKa = 4.33LINTQPKK107 pKa = 10.66DD108 pKa = 3.66CFKK111 pKa = 10.31KK112 pKa = 9.82QPYY115 pKa = 7.92TVEE118 pKa = 3.49VWFNNDD124 pKa = 2.9RR125 pKa = 11.84QNSNQYY131 pKa = 10.38INWNFIYY138 pKa = 10.69YY139 pKa = 9.94QDD141 pKa = 5.44ANDD144 pKa = 3.26VWHH147 pKa = 6.85KK148 pKa = 10.54VQGLVDD154 pKa = 4.04YY155 pKa = 11.1NGLYY159 pKa = 10.1YY160 pKa = 10.63VEE162 pKa = 4.44TGGDD166 pKa = 3.16QVYY169 pKa = 9.21FALFDD174 pKa = 4.75ADD176 pKa = 3.73DD177 pKa = 3.56QRR179 pKa = 11.84FGGTGIWSVHH189 pKa = 5.39FKK191 pKa = 11.1NKK193 pKa = 8.68TLSAPSSSASSFSPHH208 pKa = 6.37SGKK211 pKa = 10.35NYY213 pKa = 9.01ATVSSPTEE221 pKa = 3.78NTVSQSEE228 pKa = 4.65SPGRR232 pKa = 11.84LQKK235 pKa = 10.67SEE237 pKa = 4.37VGSSTSPRR245 pKa = 11.84AKK247 pKa = 8.9TSTVRR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84EE256 pKa = 3.82RR257 pKa = 11.84EE258 pKa = 3.94STPEE262 pKa = 4.03GQPSTSTKK270 pKa = 9.59RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84GRR275 pKa = 11.84IGGGGGGGAEE285 pKa = 3.79RR286 pKa = 11.84AVPSPDD292 pKa = 3.2QVGSRR297 pKa = 11.84HH298 pKa = 5.85RR299 pKa = 11.84SVSTTHH305 pKa = 7.07LSRR308 pKa = 11.84LARR311 pKa = 11.84LQKK314 pKa = 10.09EE315 pKa = 4.0AWDD318 pKa = 4.01PPVLILTGPANVLKK332 pKa = 10.3CWRR335 pKa = 11.84YY336 pKa = 9.04RR337 pKa = 11.84KK338 pKa = 9.86KK339 pKa = 10.58NQNEE343 pKa = 3.9FSFLDD348 pKa = 3.16ISTIFTWVGSNKK360 pKa = 8.62EE361 pKa = 4.04TDD363 pKa = 3.17NKK365 pKa = 10.5GRR367 pKa = 11.84MLIAFHH373 pKa = 7.08NDD375 pKa = 2.94TEE377 pKa = 4.37RR378 pKa = 11.84EE379 pKa = 4.0QFVKK383 pKa = 10.33HH384 pKa = 4.43VHH386 pKa = 5.63FPKK389 pKa = 9.1HH390 pKa = 4.29TSYY393 pKa = 11.82AFGKK397 pKa = 10.11LDD399 pKa = 3.46KK400 pKa = 11.09LL401 pKa = 3.98
MM1 pKa = 8.0DD2 pKa = 4.22WLTDD6 pKa = 3.53RR7 pKa = 11.84FNAIQDD13 pKa = 3.9NILNLIEE20 pKa = 4.79QGAEE24 pKa = 4.02DD25 pKa = 5.43LDD27 pKa = 4.07SQINYY32 pKa = 7.96WNLVRR37 pKa = 11.84QEE39 pKa = 4.11NVYY42 pKa = 10.55LYY44 pKa = 10.07YY45 pKa = 10.7GRR47 pKa = 11.84KK48 pKa = 9.39EE49 pKa = 4.09GLTHH53 pKa = 7.6FGLQPLPVAAVSEE66 pKa = 4.6YY67 pKa = 10.2KK68 pKa = 10.7AKK70 pKa = 10.48EE71 pKa = 4.24AINMVLLLQSLKK83 pKa = 10.58KK84 pKa = 9.89SAYY87 pKa = 9.85ANEE90 pKa = 3.82VWTLQDD96 pKa = 4.19ASAEE100 pKa = 4.33LINTQPKK107 pKa = 10.66DD108 pKa = 3.66CFKK111 pKa = 10.31KK112 pKa = 9.82QPYY115 pKa = 7.92TVEE118 pKa = 3.49VWFNNDD124 pKa = 2.9RR125 pKa = 11.84QNSNQYY131 pKa = 10.38INWNFIYY138 pKa = 10.69YY139 pKa = 9.94QDD141 pKa = 5.44ANDD144 pKa = 3.26VWHH147 pKa = 6.85KK148 pKa = 10.54VQGLVDD154 pKa = 4.04YY155 pKa = 11.1NGLYY159 pKa = 10.1YY160 pKa = 10.63VEE162 pKa = 4.44TGGDD166 pKa = 3.16QVYY169 pKa = 9.21FALFDD174 pKa = 4.75ADD176 pKa = 3.73DD177 pKa = 3.56QRR179 pKa = 11.84FGGTGIWSVHH189 pKa = 5.39FKK191 pKa = 11.1NKK193 pKa = 8.68TLSAPSSSASSFSPHH208 pKa = 6.37SGKK211 pKa = 10.35NYY213 pKa = 9.01ATVSSPTEE221 pKa = 3.78NTVSQSEE228 pKa = 4.65SPGRR232 pKa = 11.84LQKK235 pKa = 10.67SEE237 pKa = 4.37VGSSTSPRR245 pKa = 11.84AKK247 pKa = 8.9TSTVRR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84EE256 pKa = 3.82RR257 pKa = 11.84EE258 pKa = 3.94STPEE262 pKa = 4.03GQPSTSTKK270 pKa = 9.59RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84GRR275 pKa = 11.84IGGGGGGGAEE285 pKa = 3.79RR286 pKa = 11.84AVPSPDD292 pKa = 3.2QVGSRR297 pKa = 11.84HH298 pKa = 5.85RR299 pKa = 11.84SVSTTHH305 pKa = 7.07LSRR308 pKa = 11.84LARR311 pKa = 11.84LQKK314 pKa = 10.09EE315 pKa = 4.0AWDD318 pKa = 4.01PPVLILTGPANVLKK332 pKa = 10.3CWRR335 pKa = 11.84YY336 pKa = 9.04RR337 pKa = 11.84KK338 pKa = 9.86KK339 pKa = 10.58NQNEE343 pKa = 3.9FSFLDD348 pKa = 3.16ISTIFTWVGSNKK360 pKa = 8.62EE361 pKa = 4.04TDD363 pKa = 3.17NKK365 pKa = 10.5GRR367 pKa = 11.84MLIAFHH373 pKa = 7.08NDD375 pKa = 2.94TEE377 pKa = 4.37RR378 pKa = 11.84EE379 pKa = 4.0QFVKK383 pKa = 10.33HH384 pKa = 4.43VHH386 pKa = 5.63FPKK389 pKa = 9.1HH390 pKa = 4.29TSYY393 pKa = 11.82AFGKK397 pKa = 10.11LDD399 pKa = 3.46KK400 pKa = 11.09LL401 pKa = 3.98
Molecular weight: 45.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2450 |
95 |
608 |
350.0 |
39.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.959 ± 0.288 | 2.286 ± 0.757 |
6.776 ± 0.548 | 6.0 ± 0.556 |
4.898 ± 0.41 | 5.429 ± 0.758 |
1.714 ± 0.187 | 5.429 ± 0.76 |
5.347 ± 0.812 | 8.49 ± 0.577 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.388 ± 0.294 | 4.571 ± 0.597 |
6.082 ± 0.97 | 5.429 ± 0.347 |
5.714 ± 0.709 | 6.816 ± 0.648 |
6.245 ± 0.42 | 6.612 ± 0.466 |
1.306 ± 0.33 | 3.51 ± 0.475 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
