
Ethanoligenens harbinense (strain DSM 18485 / JCM 12961 / CGMCC 1.5033 / YUAN-3)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Ethanoligenens; Ethanoligenens harbinense
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
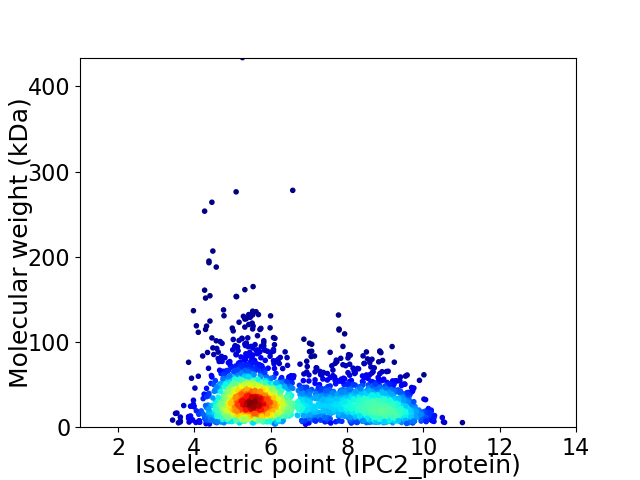
Virtual 2D-PAGE plot for 2663 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E6U6M0|E6U6M0_ETHHY Nucleoid-associated protein Ethha_0166 OS=Ethanoligenens harbinense (strain DSM 18485 / JCM 12961 / CGMCC 1.5033 / YUAN-3) OX=663278 GN=Ethha_0166 PE=3 SV=1
MM1 pKa = 7.69ASLNTGLNTALSGLSASQTALGVTAHH27 pKa = 6.72NISNAATDD35 pKa = 4.71GYY37 pKa = 9.79SRR39 pKa = 11.84QMVDD43 pKa = 3.37LQAVNVNTGAYY54 pKa = 6.73QWKK57 pKa = 8.92PVVPFVGNGVNSNQITQARR76 pKa = 11.84DD77 pKa = 2.77GFLDD81 pKa = 3.29VRR83 pKa = 11.84YY84 pKa = 8.16RR85 pKa = 11.84TANAQYY91 pKa = 10.66SDD93 pKa = 3.74YY94 pKa = 10.83EE95 pKa = 3.99QRR97 pKa = 11.84QNDD100 pKa = 3.77LGQVEE105 pKa = 5.61DD106 pKa = 4.04IFNEE110 pKa = 4.13VSTSGNDD117 pKa = 3.47TLQGLSGQLNTLINDD132 pKa = 3.35IGSYY136 pKa = 9.63QSSPASSSLPVTIKK150 pKa = 9.93TDD152 pKa = 3.39AEE154 pKa = 4.11NLVTKK159 pKa = 10.47IRR161 pKa = 11.84TAYY164 pKa = 10.53SSLTTFEE171 pKa = 4.33SQEE174 pKa = 3.8QSALNVVVSGDD185 pKa = 3.78TQSGGINGILQNISTLNTQIVSMEE209 pKa = 4.25VGGQPANDD217 pKa = 4.01LRR219 pKa = 11.84DD220 pKa = 3.49QRR222 pKa = 11.84NQLLDD227 pKa = 3.61KK228 pKa = 11.09LSGYY232 pKa = 10.68LDD234 pKa = 3.54ISATEE239 pKa = 3.87QSDD242 pKa = 3.89GSVTVQMEE250 pKa = 3.91NDD252 pKa = 3.61LASGSGAMLVDD263 pKa = 4.19GNNIVHH269 pKa = 5.9TLKK272 pKa = 10.85VVTDD276 pKa = 4.69GPTDD280 pKa = 3.53ASGLSTTAVTWDD292 pKa = 3.71ASDD295 pKa = 4.49SNNQSTTYY303 pKa = 9.25ATNTGTGWSGNTLSVGGGSVNGYY326 pKa = 9.38LAILNGDD333 pKa = 3.55GSGTGSYY340 pKa = 11.07GDD342 pKa = 3.68VGVHH346 pKa = 5.48YY347 pKa = 10.53LKK349 pKa = 10.89EE350 pKa = 4.07HH351 pKa = 6.63LNDD354 pKa = 4.04FAQSFADD361 pKa = 3.43IMNSTSTSSGGGAQMLTYY379 pKa = 10.79GSGTPPTPTYY389 pKa = 10.43PDD391 pKa = 3.3TTGWSDD397 pKa = 3.39PTASAAYY404 pKa = 9.65VATDD408 pKa = 3.71AAATISLSDD417 pKa = 3.6AWNKK421 pKa = 10.99DD422 pKa = 3.04DD423 pKa = 5.54TLFGDD428 pKa = 4.17NYY430 pKa = 9.8TGTSVGTYY438 pKa = 9.96AQAFLNALGSAQVSSYY454 pKa = 9.6DD455 pKa = 3.56TSAHH459 pKa = 5.37KK460 pKa = 9.05TLYY463 pKa = 10.1TDD465 pKa = 3.25GTMASYY471 pKa = 10.87AATFSNKK478 pKa = 8.6IANVINDD485 pKa = 4.15DD486 pKa = 3.21TTMAGNYY493 pKa = 9.09QIQKK497 pKa = 10.31NDD499 pKa = 4.02LDD501 pKa = 3.95TQRR504 pKa = 11.84QSVSSVSIDD513 pKa = 3.49EE514 pKa = 4.26EE515 pKa = 4.8TVNLMKK521 pKa = 10.38FQQMYY526 pKa = 8.93GASARR531 pKa = 11.84VITTINDD538 pKa = 3.25MMGTLLSMAQQ548 pKa = 3.16
MM1 pKa = 7.69ASLNTGLNTALSGLSASQTALGVTAHH27 pKa = 6.72NISNAATDD35 pKa = 4.71GYY37 pKa = 9.79SRR39 pKa = 11.84QMVDD43 pKa = 3.37LQAVNVNTGAYY54 pKa = 6.73QWKK57 pKa = 8.92PVVPFVGNGVNSNQITQARR76 pKa = 11.84DD77 pKa = 2.77GFLDD81 pKa = 3.29VRR83 pKa = 11.84YY84 pKa = 8.16RR85 pKa = 11.84TANAQYY91 pKa = 10.66SDD93 pKa = 3.74YY94 pKa = 10.83EE95 pKa = 3.99QRR97 pKa = 11.84QNDD100 pKa = 3.77LGQVEE105 pKa = 5.61DD106 pKa = 4.04IFNEE110 pKa = 4.13VSTSGNDD117 pKa = 3.47TLQGLSGQLNTLINDD132 pKa = 3.35IGSYY136 pKa = 9.63QSSPASSSLPVTIKK150 pKa = 9.93TDD152 pKa = 3.39AEE154 pKa = 4.11NLVTKK159 pKa = 10.47IRR161 pKa = 11.84TAYY164 pKa = 10.53SSLTTFEE171 pKa = 4.33SQEE174 pKa = 3.8QSALNVVVSGDD185 pKa = 3.78TQSGGINGILQNISTLNTQIVSMEE209 pKa = 4.25VGGQPANDD217 pKa = 4.01LRR219 pKa = 11.84DD220 pKa = 3.49QRR222 pKa = 11.84NQLLDD227 pKa = 3.61KK228 pKa = 11.09LSGYY232 pKa = 10.68LDD234 pKa = 3.54ISATEE239 pKa = 3.87QSDD242 pKa = 3.89GSVTVQMEE250 pKa = 3.91NDD252 pKa = 3.61LASGSGAMLVDD263 pKa = 4.19GNNIVHH269 pKa = 5.9TLKK272 pKa = 10.85VVTDD276 pKa = 4.69GPTDD280 pKa = 3.53ASGLSTTAVTWDD292 pKa = 3.71ASDD295 pKa = 4.49SNNQSTTYY303 pKa = 9.25ATNTGTGWSGNTLSVGGGSVNGYY326 pKa = 9.38LAILNGDD333 pKa = 3.55GSGTGSYY340 pKa = 11.07GDD342 pKa = 3.68VGVHH346 pKa = 5.48YY347 pKa = 10.53LKK349 pKa = 10.89EE350 pKa = 4.07HH351 pKa = 6.63LNDD354 pKa = 4.04FAQSFADD361 pKa = 3.43IMNSTSTSSGGGAQMLTYY379 pKa = 10.79GSGTPPTPTYY389 pKa = 10.43PDD391 pKa = 3.3TTGWSDD397 pKa = 3.39PTASAAYY404 pKa = 9.65VATDD408 pKa = 3.71AAATISLSDD417 pKa = 3.6AWNKK421 pKa = 10.99DD422 pKa = 3.04DD423 pKa = 5.54TLFGDD428 pKa = 4.17NYY430 pKa = 9.8TGTSVGTYY438 pKa = 9.96AQAFLNALGSAQVSSYY454 pKa = 9.6DD455 pKa = 3.56TSAHH459 pKa = 5.37KK460 pKa = 9.05TLYY463 pKa = 10.1TDD465 pKa = 3.25GTMASYY471 pKa = 10.87AATFSNKK478 pKa = 8.6IANVINDD485 pKa = 4.15DD486 pKa = 3.21TTMAGNYY493 pKa = 9.09QIQKK497 pKa = 10.31NDD499 pKa = 4.02LDD501 pKa = 3.95TQRR504 pKa = 11.84QSVSSVSIDD513 pKa = 3.49EE514 pKa = 4.26EE515 pKa = 4.8TVNLMKK521 pKa = 10.38FQQMYY526 pKa = 8.93GASARR531 pKa = 11.84VITTINDD538 pKa = 3.25MMGTLLSMAQQ548 pKa = 3.16
Molecular weight: 57.49 kDa
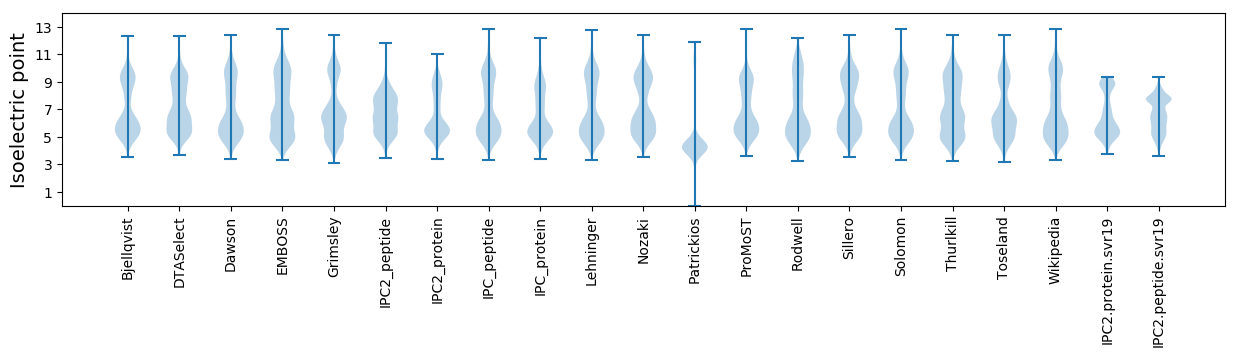
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E6U542|E6U542_ETHHY DUF2344 domain-containing protein OS=Ethanoligenens harbinense (strain DSM 18485 / JCM 12961 / CGMCC 1.5033 / YUAN-3) OX=663278 GN=Ethha_1197 PE=4 SV=1
MM1 pKa = 7.62IKK3 pKa = 9.2TPRR6 pKa = 11.84NQPPRR11 pKa = 11.84IKK13 pKa = 10.39EE14 pKa = 3.84RR15 pKa = 11.84LHH17 pKa = 6.89AFLLTSGASFLLLLLLFSPQAGLDD41 pKa = 3.64GARR44 pKa = 11.84TGLLLCAEE52 pKa = 4.49VVIPTLFPFLVLSSFVIHH70 pKa = 6.87LGLAEE75 pKa = 3.94RR76 pKa = 11.84FGRR79 pKa = 11.84LFTRR83 pKa = 11.84LTAALFKK90 pKa = 11.05LPGASAPVLVLGILGGYY107 pKa = 8.5PVGARR112 pKa = 11.84ACAEE116 pKa = 4.03LVDD119 pKa = 4.16TGALTRR125 pKa = 11.84AEE127 pKa = 4.36GNRR130 pKa = 11.84LLTFCINSSPAYY142 pKa = 9.73IIGAAGAGLLHH153 pKa = 6.73SAQAGALLYY162 pKa = 10.12AAHH165 pKa = 7.11ILSSLLVGTLLGMHH179 pKa = 6.26SPKK182 pKa = 10.29PSAGALARR190 pKa = 11.84RR191 pKa = 11.84PVTRR195 pKa = 11.84MPASGALVTSVTGAATSMLGISGFVVFFSSLSSLLFSTGLVGWMARR241 pKa = 11.84LAAFLPGLHH250 pKa = 6.28WFDD253 pKa = 4.59AGTAQTLLKK262 pKa = 10.81GFLEE266 pKa = 4.45VSGGCDD272 pKa = 2.69AAVHH276 pKa = 6.1NGQFVLEE283 pKa = 4.24ALSAFLSWSGLSVVFQVLFAVRR305 pKa = 11.84GTGLSARR312 pKa = 11.84GYY314 pKa = 8.05VASRR318 pKa = 11.84PLHH321 pKa = 5.31MLFSILFTVILFSLFPVAIPTLAATSRR348 pKa = 11.84VALSMHH354 pKa = 6.55NAPASAALLFACALLLLARR373 pKa = 11.84ATVV376 pKa = 3.18
MM1 pKa = 7.62IKK3 pKa = 9.2TPRR6 pKa = 11.84NQPPRR11 pKa = 11.84IKK13 pKa = 10.39EE14 pKa = 3.84RR15 pKa = 11.84LHH17 pKa = 6.89AFLLTSGASFLLLLLLFSPQAGLDD41 pKa = 3.64GARR44 pKa = 11.84TGLLLCAEE52 pKa = 4.49VVIPTLFPFLVLSSFVIHH70 pKa = 6.87LGLAEE75 pKa = 3.94RR76 pKa = 11.84FGRR79 pKa = 11.84LFTRR83 pKa = 11.84LTAALFKK90 pKa = 11.05LPGASAPVLVLGILGGYY107 pKa = 8.5PVGARR112 pKa = 11.84ACAEE116 pKa = 4.03LVDD119 pKa = 4.16TGALTRR125 pKa = 11.84AEE127 pKa = 4.36GNRR130 pKa = 11.84LLTFCINSSPAYY142 pKa = 9.73IIGAAGAGLLHH153 pKa = 6.73SAQAGALLYY162 pKa = 10.12AAHH165 pKa = 7.11ILSSLLVGTLLGMHH179 pKa = 6.26SPKK182 pKa = 10.29PSAGALARR190 pKa = 11.84RR191 pKa = 11.84PVTRR195 pKa = 11.84MPASGALVTSVTGAATSMLGISGFVVFFSSLSSLLFSTGLVGWMARR241 pKa = 11.84LAAFLPGLHH250 pKa = 6.28WFDD253 pKa = 4.59AGTAQTLLKK262 pKa = 10.81GFLEE266 pKa = 4.45VSGGCDD272 pKa = 2.69AAVHH276 pKa = 6.1NGQFVLEE283 pKa = 4.24ALSAFLSWSGLSVVFQVLFAVRR305 pKa = 11.84GTGLSARR312 pKa = 11.84GYY314 pKa = 8.05VASRR318 pKa = 11.84PLHH321 pKa = 5.31MLFSILFTVILFSLFPVAIPTLAATSRR348 pKa = 11.84VALSMHH354 pKa = 6.55NAPASAALLFACALLLLARR373 pKa = 11.84ATVV376 pKa = 3.18
Molecular weight: 39.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
838389 |
31 |
3906 |
314.8 |
34.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.012 ± 0.064 | 1.397 ± 0.021 |
5.457 ± 0.039 | 5.829 ± 0.062 |
4.095 ± 0.032 | 7.801 ± 0.044 |
2.01 ± 0.021 | 5.972 ± 0.049 |
4.944 ± 0.042 | 9.705 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.022 | 3.674 ± 0.04 |
4.119 ± 0.028 | 3.567 ± 0.033 |
5.566 ± 0.062 | 5.97 ± 0.069 |
5.773 ± 0.062 | 7.315 ± 0.042 |
0.913 ± 0.014 | 3.349 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
