
Butyrivibrio sp. INlla14
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Butyrivibrio; unclassified Butyrivibrio
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
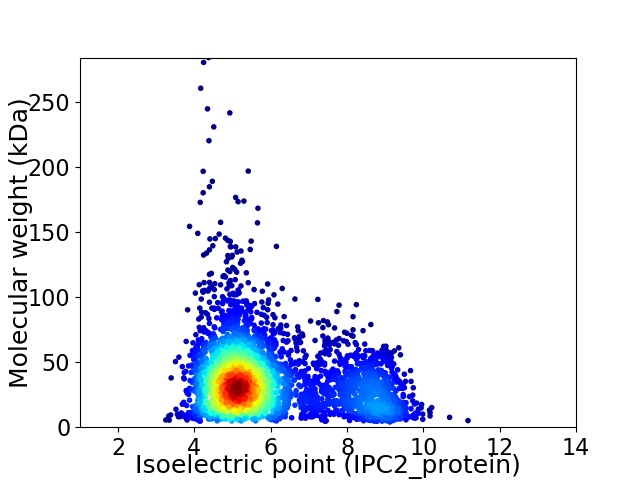
Virtual 2D-PAGE plot for 3697 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G5H9Y9|A0A1G5H9Y9_9FIRM Membrane protein implicated in regulation of membrane protease activity OS=Butyrivibrio sp. INlla14 OX=1520808 GN=SAMN02910371_02991 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 10.44KK3 pKa = 10.27KK4 pKa = 10.27IAVLLAGVMVCTMFTACGKK23 pKa = 10.13DD24 pKa = 3.27AGSDD28 pKa = 3.4AANEE32 pKa = 4.44AISVNEE38 pKa = 3.87EE39 pKa = 3.85ALASAAADD47 pKa = 3.31VSTEE51 pKa = 4.05GEE53 pKa = 4.32DD54 pKa = 3.16TGLTIDD60 pKa = 4.63FDD62 pKa = 4.17NLEE65 pKa = 4.25TSTLQDD71 pKa = 2.8IKK73 pKa = 11.33ASDD76 pKa = 3.91LVTLGEE82 pKa = 4.33YY83 pKa = 10.65NGITVEE89 pKa = 4.06ATLGEE94 pKa = 4.34VTEE97 pKa = 4.53EE98 pKa = 4.04DD99 pKa = 3.58VAGYY103 pKa = 9.47IEE105 pKa = 4.63NMKK108 pKa = 10.51SSNPPMIDD116 pKa = 3.03VTDD119 pKa = 3.99RR120 pKa = 11.84AVQDD124 pKa = 3.49GDD126 pKa = 4.11TVNIDD131 pKa = 3.63YY132 pKa = 10.54VGKK135 pKa = 10.53YY136 pKa = 10.47ADD138 pKa = 3.68TKK140 pKa = 10.45EE141 pKa = 4.28AFDD144 pKa = 4.2GGTANGADD152 pKa = 3.94LVIGSNSYY160 pKa = 10.54IEE162 pKa = 4.6GFEE165 pKa = 4.16SGLVGAEE172 pKa = 3.58IGEE175 pKa = 4.34TRR177 pKa = 11.84DD178 pKa = 3.75LNLTFPADD186 pKa = 3.58YY187 pKa = 10.24GAEE190 pKa = 4.04SLAGKK195 pKa = 10.11DD196 pKa = 3.24VVFTVTINSIKK207 pKa = 10.38VAADD211 pKa = 4.34DD212 pKa = 5.99ISDD215 pKa = 3.27EE216 pKa = 4.09WAAGLGIADD225 pKa = 3.58VTTISEE231 pKa = 4.27LEE233 pKa = 4.02AYY235 pKa = 9.85ANKK238 pKa = 9.44TLTTQAEE245 pKa = 4.31DD246 pKa = 3.96TYY248 pKa = 11.45KK249 pKa = 10.1STIEE253 pKa = 3.9NSAVQQVYY261 pKa = 10.34DD262 pKa = 3.47SCAFGDD268 pKa = 4.07IPQTLINRR276 pKa = 11.84YY277 pKa = 9.26LKK279 pKa = 10.4LQKK282 pKa = 10.65QMLDD286 pKa = 3.68YY287 pKa = 10.54QATMYY292 pKa = 10.38SYY294 pKa = 11.6YY295 pKa = 10.67YY296 pKa = 9.88GQQLSASDD304 pKa = 4.88LISVYY309 pKa = 10.09MNNEE313 pKa = 3.86GFVGTADD320 pKa = 5.56DD321 pKa = 4.24YY322 pKa = 11.5LQSISTDD329 pKa = 3.09MAKK332 pKa = 10.08QYY334 pKa = 11.68LMFQAIADD342 pKa = 3.99EE343 pKa = 4.07QGITISDD350 pKa = 3.83EE351 pKa = 5.77DD352 pKa = 3.87IDD354 pKa = 4.52TYY356 pKa = 11.62LKK358 pKa = 10.62DD359 pKa = 3.53AYY361 pKa = 9.68EE362 pKa = 4.36NASTTSFSSYY372 pKa = 10.65EE373 pKa = 4.02EE374 pKa = 4.18YY375 pKa = 10.47KK376 pKa = 11.0ASLDD380 pKa = 3.56LEE382 pKa = 4.7TYY384 pKa = 10.47RR385 pKa = 11.84EE386 pKa = 3.83GLMADD391 pKa = 3.5KK392 pKa = 10.67AVEE395 pKa = 4.67FIVNNANVVAAPVEE409 pKa = 4.4ATTEE413 pKa = 4.12TSAEE417 pKa = 4.05ASSEE421 pKa = 4.05ASVDD425 pKa = 3.35AAAEE429 pKa = 4.15SSSTVAEE436 pKa = 4.2
MM1 pKa = 7.37KK2 pKa = 10.44KK3 pKa = 10.27KK4 pKa = 10.27IAVLLAGVMVCTMFTACGKK23 pKa = 10.13DD24 pKa = 3.27AGSDD28 pKa = 3.4AANEE32 pKa = 4.44AISVNEE38 pKa = 3.87EE39 pKa = 3.85ALASAAADD47 pKa = 3.31VSTEE51 pKa = 4.05GEE53 pKa = 4.32DD54 pKa = 3.16TGLTIDD60 pKa = 4.63FDD62 pKa = 4.17NLEE65 pKa = 4.25TSTLQDD71 pKa = 2.8IKK73 pKa = 11.33ASDD76 pKa = 3.91LVTLGEE82 pKa = 4.33YY83 pKa = 10.65NGITVEE89 pKa = 4.06ATLGEE94 pKa = 4.34VTEE97 pKa = 4.53EE98 pKa = 4.04DD99 pKa = 3.58VAGYY103 pKa = 9.47IEE105 pKa = 4.63NMKK108 pKa = 10.51SSNPPMIDD116 pKa = 3.03VTDD119 pKa = 3.99RR120 pKa = 11.84AVQDD124 pKa = 3.49GDD126 pKa = 4.11TVNIDD131 pKa = 3.63YY132 pKa = 10.54VGKK135 pKa = 10.53YY136 pKa = 10.47ADD138 pKa = 3.68TKK140 pKa = 10.45EE141 pKa = 4.28AFDD144 pKa = 4.2GGTANGADD152 pKa = 3.94LVIGSNSYY160 pKa = 10.54IEE162 pKa = 4.6GFEE165 pKa = 4.16SGLVGAEE172 pKa = 3.58IGEE175 pKa = 4.34TRR177 pKa = 11.84DD178 pKa = 3.75LNLTFPADD186 pKa = 3.58YY187 pKa = 10.24GAEE190 pKa = 4.04SLAGKK195 pKa = 10.11DD196 pKa = 3.24VVFTVTINSIKK207 pKa = 10.38VAADD211 pKa = 4.34DD212 pKa = 5.99ISDD215 pKa = 3.27EE216 pKa = 4.09WAAGLGIADD225 pKa = 3.58VTTISEE231 pKa = 4.27LEE233 pKa = 4.02AYY235 pKa = 9.85ANKK238 pKa = 9.44TLTTQAEE245 pKa = 4.31DD246 pKa = 3.96TYY248 pKa = 11.45KK249 pKa = 10.1STIEE253 pKa = 3.9NSAVQQVYY261 pKa = 10.34DD262 pKa = 3.47SCAFGDD268 pKa = 4.07IPQTLINRR276 pKa = 11.84YY277 pKa = 9.26LKK279 pKa = 10.4LQKK282 pKa = 10.65QMLDD286 pKa = 3.68YY287 pKa = 10.54QATMYY292 pKa = 10.38SYY294 pKa = 11.6YY295 pKa = 10.67YY296 pKa = 9.88GQQLSASDD304 pKa = 4.88LISVYY309 pKa = 10.09MNNEE313 pKa = 3.86GFVGTADD320 pKa = 5.56DD321 pKa = 4.24YY322 pKa = 11.5LQSISTDD329 pKa = 3.09MAKK332 pKa = 10.08QYY334 pKa = 11.68LMFQAIADD342 pKa = 3.99EE343 pKa = 4.07QGITISDD350 pKa = 3.83EE351 pKa = 5.77DD352 pKa = 3.87IDD354 pKa = 4.52TYY356 pKa = 11.62LKK358 pKa = 10.62DD359 pKa = 3.53AYY361 pKa = 9.68EE362 pKa = 4.36NASTTSFSSYY372 pKa = 10.65EE373 pKa = 4.02EE374 pKa = 4.18YY375 pKa = 10.47KK376 pKa = 11.0ASLDD380 pKa = 3.56LEE382 pKa = 4.7TYY384 pKa = 10.47RR385 pKa = 11.84EE386 pKa = 3.83GLMADD391 pKa = 3.5KK392 pKa = 10.67AVEE395 pKa = 4.67FIVNNANVVAAPVEE409 pKa = 4.4ATTEE413 pKa = 4.12TSAEE417 pKa = 4.05ASSEE421 pKa = 4.05ASVDD425 pKa = 3.35AAAEE429 pKa = 4.15SSSTVAEE436 pKa = 4.2
Molecular weight: 46.56 kDa
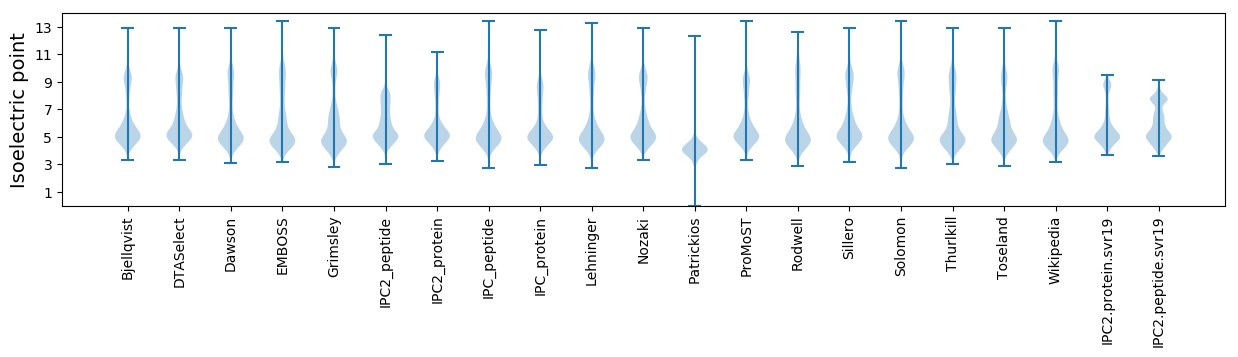
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G5BRX8|A0A1G5BRX8_9FIRM SCP-2 sterol transfer family protein OS=Butyrivibrio sp. INlla14 OX=1520808 GN=SAMN02910371_00473 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 8.74MTFQPHH8 pKa = 5.93KK9 pKa = 8.68LQRR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.36GGRR28 pKa = 11.84KK29 pKa = 8.97VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.58KK2 pKa = 8.74MTFQPHH8 pKa = 5.93KK9 pKa = 8.68LQRR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.36GGRR28 pKa = 11.84KK29 pKa = 8.97VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1242442 |
39 |
2640 |
336.1 |
37.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.288 ± 0.041 | 1.343 ± 0.015 |
6.517 ± 0.029 | 7.333 ± 0.042 |
4.357 ± 0.029 | 6.88 ± 0.04 |
1.625 ± 0.017 | 7.6 ± 0.038 |
7.096 ± 0.034 | 8.437 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.994 ± 0.019 | 4.79 ± 0.028 |
3.093 ± 0.022 | 2.815 ± 0.021 |
4.077 ± 0.029 | 6.234 ± 0.035 |
5.308 ± 0.038 | 6.815 ± 0.034 |
0.931 ± 0.013 | 4.467 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
