
Llama faeces associated circular DNA virus-1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.55
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
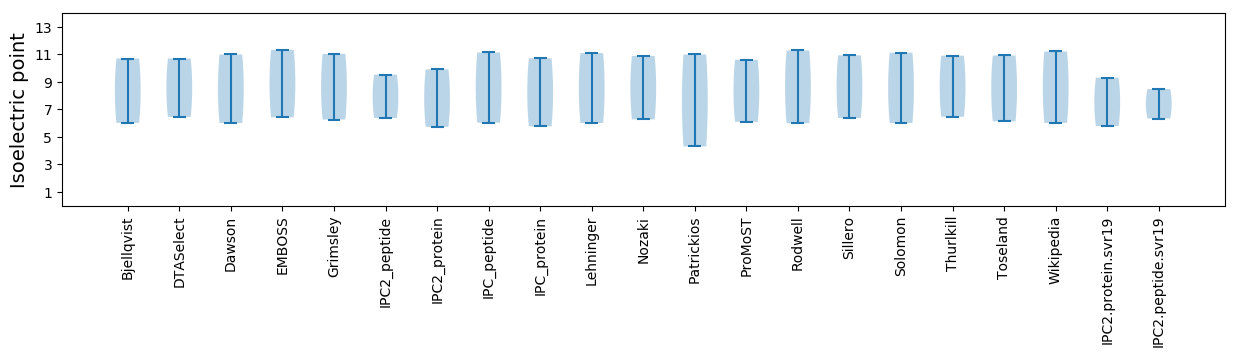
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160HWJ8|A0A160HWJ8_9VIRU Putative capsid protein OS=Llama faeces associated circular DNA virus-1 OX=1843771 PE=4 SV=1
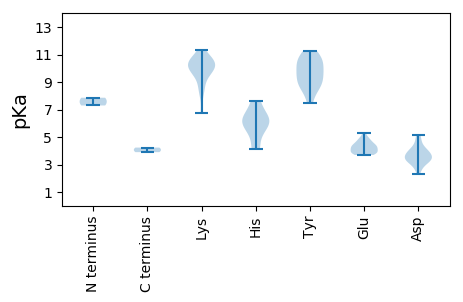
MM1 pKa = 7.86PSNPQFFHH9 pKa = 7.65FCFTLNNYY17 pKa = 8.62VEE19 pKa = 4.5EE20 pKa = 4.22VLDD23 pKa = 4.06DD24 pKa = 3.97VEE26 pKa = 4.93EE27 pKa = 4.32EE28 pKa = 4.1DD29 pKa = 4.27VPRR32 pKa = 11.84ISAFCEE38 pKa = 3.89EE39 pKa = 4.43EE40 pKa = 3.81GKK42 pKa = 10.62YY43 pKa = 9.68WIIGRR48 pKa = 11.84EE49 pKa = 4.18VGEE52 pKa = 4.35SGTPHH57 pKa = 5.68LQGYY61 pKa = 9.13VSLRR65 pKa = 11.84KK66 pKa = 9.36RR67 pKa = 11.84RR68 pKa = 11.84TFVYY72 pKa = 10.61VRR74 pKa = 11.84DD75 pKa = 3.82KK76 pKa = 11.33LSNRR80 pKa = 11.84CHH82 pKa = 6.55IEE84 pKa = 3.7SSRR87 pKa = 11.84GTARR91 pKa = 11.84QNRR94 pKa = 11.84EE95 pKa = 3.81YY96 pKa = 10.52CSKK99 pKa = 10.71GGNFIEE105 pKa = 4.94GGSINEE111 pKa = 3.69GRR113 pKa = 11.84VRR115 pKa = 11.84QDD117 pKa = 2.34RR118 pKa = 11.84DD119 pKa = 2.82EE120 pKa = 4.26AARR123 pKa = 11.84SFMAAVRR130 pKa = 11.84RR131 pKa = 11.84GDD133 pKa = 3.27QGLVEE138 pKa = 4.45FADD141 pKa = 4.29AEE143 pKa = 4.05PHH145 pKa = 4.11TWIRR149 pKa = 11.84HH150 pKa = 4.19GSNMLRR156 pKa = 11.84NALSILPPIEE166 pKa = 4.3RR167 pKa = 11.84PSISVRR173 pKa = 11.84WIWGSPGVGKK183 pKa = 10.57SRR185 pKa = 11.84IAHH188 pKa = 5.28EE189 pKa = 4.58TLPDD193 pKa = 3.72AYY195 pKa = 11.22VKK197 pKa = 10.61DD198 pKa = 4.61PRR200 pKa = 11.84TKK202 pKa = 8.07WWNGYY207 pKa = 8.98LCQEE211 pKa = 4.71DD212 pKa = 5.12VIIDD216 pKa = 3.66DD217 pKa = 4.93FGPNGIDD224 pKa = 3.32INHH227 pKa = 7.46LLRR230 pKa = 11.84WFDD233 pKa = 3.45RR234 pKa = 11.84YY235 pKa = 10.77KK236 pKa = 11.0CLVEE240 pKa = 4.28NKK242 pKa = 9.85GGMVALHH249 pKa = 5.86ATNFIVTSNFHH260 pKa = 6.34PRR262 pKa = 11.84DD263 pKa = 3.08IFKK266 pKa = 10.81FGDD269 pKa = 3.95EE270 pKa = 4.81INPQLPALEE279 pKa = 3.79RR280 pKa = 11.84RR281 pKa = 11.84IVIEE285 pKa = 3.93EE286 pKa = 3.96MLL288 pKa = 4.23
MM1 pKa = 7.86PSNPQFFHH9 pKa = 7.65FCFTLNNYY17 pKa = 8.62VEE19 pKa = 4.5EE20 pKa = 4.22VLDD23 pKa = 4.06DD24 pKa = 3.97VEE26 pKa = 4.93EE27 pKa = 4.32EE28 pKa = 4.1DD29 pKa = 4.27VPRR32 pKa = 11.84ISAFCEE38 pKa = 3.89EE39 pKa = 4.43EE40 pKa = 3.81GKK42 pKa = 10.62YY43 pKa = 9.68WIIGRR48 pKa = 11.84EE49 pKa = 4.18VGEE52 pKa = 4.35SGTPHH57 pKa = 5.68LQGYY61 pKa = 9.13VSLRR65 pKa = 11.84KK66 pKa = 9.36RR67 pKa = 11.84RR68 pKa = 11.84TFVYY72 pKa = 10.61VRR74 pKa = 11.84DD75 pKa = 3.82KK76 pKa = 11.33LSNRR80 pKa = 11.84CHH82 pKa = 6.55IEE84 pKa = 3.7SSRR87 pKa = 11.84GTARR91 pKa = 11.84QNRR94 pKa = 11.84EE95 pKa = 3.81YY96 pKa = 10.52CSKK99 pKa = 10.71GGNFIEE105 pKa = 4.94GGSINEE111 pKa = 3.69GRR113 pKa = 11.84VRR115 pKa = 11.84QDD117 pKa = 2.34RR118 pKa = 11.84DD119 pKa = 2.82EE120 pKa = 4.26AARR123 pKa = 11.84SFMAAVRR130 pKa = 11.84RR131 pKa = 11.84GDD133 pKa = 3.27QGLVEE138 pKa = 4.45FADD141 pKa = 4.29AEE143 pKa = 4.05PHH145 pKa = 4.11TWIRR149 pKa = 11.84HH150 pKa = 4.19GSNMLRR156 pKa = 11.84NALSILPPIEE166 pKa = 4.3RR167 pKa = 11.84PSISVRR173 pKa = 11.84WIWGSPGVGKK183 pKa = 10.57SRR185 pKa = 11.84IAHH188 pKa = 5.28EE189 pKa = 4.58TLPDD193 pKa = 3.72AYY195 pKa = 11.22VKK197 pKa = 10.61DD198 pKa = 4.61PRR200 pKa = 11.84TKK202 pKa = 8.07WWNGYY207 pKa = 8.98LCQEE211 pKa = 4.71DD212 pKa = 5.12VIIDD216 pKa = 3.66DD217 pKa = 4.93FGPNGIDD224 pKa = 3.32INHH227 pKa = 7.46LLRR230 pKa = 11.84WFDD233 pKa = 3.45RR234 pKa = 11.84YY235 pKa = 10.77KK236 pKa = 11.0CLVEE240 pKa = 4.28NKK242 pKa = 9.85GGMVALHH249 pKa = 5.86ATNFIVTSNFHH260 pKa = 6.34PRR262 pKa = 11.84DD263 pKa = 3.08IFKK266 pKa = 10.81FGDD269 pKa = 3.95EE270 pKa = 4.81INPQLPALEE279 pKa = 3.79RR280 pKa = 11.84RR281 pKa = 11.84IVIEE285 pKa = 3.93EE286 pKa = 3.96MLL288 pKa = 4.23
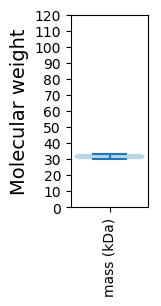
Molecular weight: 33.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160HWJ8|A0A160HWJ8_9VIRU Putative capsid protein OS=Llama faeces associated circular DNA virus-1 OX=1843771 PE=4 SV=1
MM1 pKa = 7.29AFKK4 pKa = 10.49RR5 pKa = 11.84KK6 pKa = 9.11RR7 pKa = 11.84VYY9 pKa = 10.71APRR12 pKa = 11.84ANAFKK17 pKa = 10.42KK18 pKa = 10.13RR19 pKa = 11.84KK20 pKa = 8.62SAMRR24 pKa = 11.84RR25 pKa = 11.84KK26 pKa = 9.58RR27 pKa = 11.84RR28 pKa = 11.84NSKK31 pKa = 9.82GGPRR35 pKa = 11.84VVGYY39 pKa = 8.88TSLNQKK45 pKa = 9.86DD46 pKa = 3.42HH47 pKa = 6.27TFGFRR52 pKa = 11.84ARR54 pKa = 11.84KK55 pKa = 6.74TSRR58 pKa = 11.84RR59 pKa = 11.84AFKK62 pKa = 10.46KK63 pKa = 10.1HH64 pKa = 5.74LWDD67 pKa = 3.29STLYY71 pKa = 9.69ANHH74 pKa = 6.49WRR76 pKa = 11.84SFSSSQQNVTTPAGTLDD93 pKa = 3.46TALFAINLTSPIGNTSPFWTAAGGLVPTDD122 pKa = 3.34IGVTPPKK129 pKa = 10.25FVGDD133 pKa = 3.21ITLRR137 pKa = 11.84GGVWEE142 pKa = 4.66CIIYY146 pKa = 9.78NQSTTQDD153 pKa = 2.73IRR155 pKa = 11.84FKK157 pKa = 9.68SWLVRR162 pKa = 11.84TVDD165 pKa = 3.73TPNFTPFPATSPISWEE181 pKa = 3.71PSIAADD187 pKa = 3.94FVQYY191 pKa = 10.03IGKK194 pKa = 8.49PFQSRR199 pKa = 11.84EE200 pKa = 3.87VVIEE204 pKa = 3.99SGNSYY209 pKa = 7.49TLKK212 pKa = 10.2GRR214 pKa = 11.84YY215 pKa = 8.48RR216 pKa = 11.84VEE218 pKa = 5.29KK219 pKa = 10.04IDD221 pKa = 3.72QKK223 pKa = 10.04VQAVAGRR230 pKa = 11.84QLFLFGLIHH239 pKa = 6.34NVGSAGAVAQTMNIVTSQNLSFAADD264 pKa = 3.39AVGTTT269 pKa = 3.9
MM1 pKa = 7.29AFKK4 pKa = 10.49RR5 pKa = 11.84KK6 pKa = 9.11RR7 pKa = 11.84VYY9 pKa = 10.71APRR12 pKa = 11.84ANAFKK17 pKa = 10.42KK18 pKa = 10.13RR19 pKa = 11.84KK20 pKa = 8.62SAMRR24 pKa = 11.84RR25 pKa = 11.84KK26 pKa = 9.58RR27 pKa = 11.84RR28 pKa = 11.84NSKK31 pKa = 9.82GGPRR35 pKa = 11.84VVGYY39 pKa = 8.88TSLNQKK45 pKa = 9.86DD46 pKa = 3.42HH47 pKa = 6.27TFGFRR52 pKa = 11.84ARR54 pKa = 11.84KK55 pKa = 6.74TSRR58 pKa = 11.84RR59 pKa = 11.84AFKK62 pKa = 10.46KK63 pKa = 10.1HH64 pKa = 5.74LWDD67 pKa = 3.29STLYY71 pKa = 9.69ANHH74 pKa = 6.49WRR76 pKa = 11.84SFSSSQQNVTTPAGTLDD93 pKa = 3.46TALFAINLTSPIGNTSPFWTAAGGLVPTDD122 pKa = 3.34IGVTPPKK129 pKa = 10.25FVGDD133 pKa = 3.21ITLRR137 pKa = 11.84GGVWEE142 pKa = 4.66CIIYY146 pKa = 9.78NQSTTQDD153 pKa = 2.73IRR155 pKa = 11.84FKK157 pKa = 9.68SWLVRR162 pKa = 11.84TVDD165 pKa = 3.73TPNFTPFPATSPISWEE181 pKa = 3.71PSIAADD187 pKa = 3.94FVQYY191 pKa = 10.03IGKK194 pKa = 8.49PFQSRR199 pKa = 11.84EE200 pKa = 3.87VVIEE204 pKa = 3.99SGNSYY209 pKa = 7.49TLKK212 pKa = 10.2GRR214 pKa = 11.84YY215 pKa = 8.48RR216 pKa = 11.84VEE218 pKa = 5.29KK219 pKa = 10.04IDD221 pKa = 3.72QKK223 pKa = 10.04VQAVAGRR230 pKa = 11.84QLFLFGLIHH239 pKa = 6.34NVGSAGAVAQTMNIVTSQNLSFAADD264 pKa = 3.39AVGTTT269 pKa = 3.9
Molecular weight: 29.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
557 |
269 |
288 |
278.5 |
31.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.822 ± 1.333 | 1.257 ± 0.562 |
4.847 ± 0.717 | 5.386 ± 2.239 |
5.745 ± 0.365 | 7.72 ± 0.181 |
2.334 ± 0.538 | 6.284 ± 0.685 |
4.847 ± 0.935 | 5.745 ± 0.343 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.436 ± 0.204 | 5.206 ± 0.237 |
5.206 ± 0.001 | 3.411 ± 0.666 |
8.618 ± 0.515 | 7.002 ± 0.747 |
6.104 ± 2.024 | 7.002 ± 0.275 |
2.334 ± 0.066 | 2.693 ± 0.058 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
