
Torque teno virus (isolate Chimpanzee/Japan/Pt-TTV6/2000) (TTV)
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 8.1
Get precalculated fractions of proteins
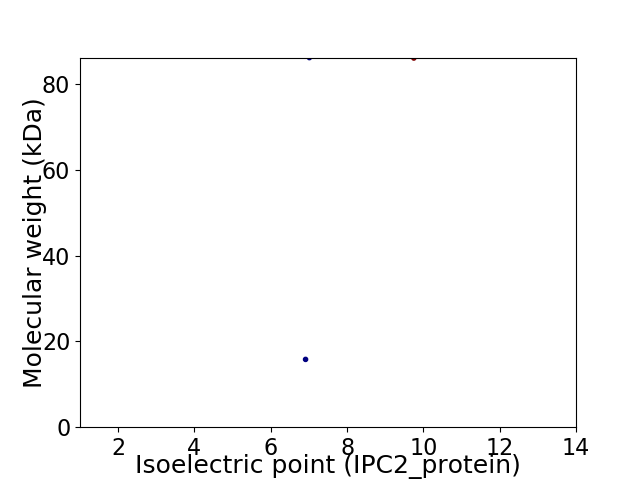
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9DUD0|ORF2_TTVV8 Uncharacterized ORF2 protein OS=Torque teno virus (isolate Chimpanzee/Japan/Pt-TTV6/2000) OX=687343 GN=ORF2 PE=4 SV=1
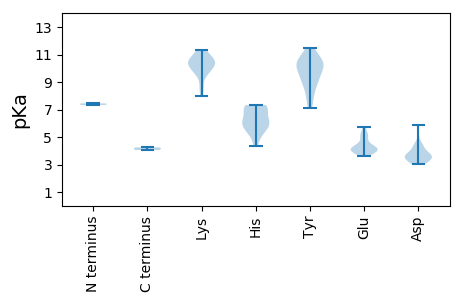
MM1 pKa = 7.45HH2 pKa = 7.29FSKK5 pKa = 10.89LSRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.51KK11 pKa = 10.58GKK13 pKa = 10.24VPLPPLPTAPLPPHH27 pKa = 7.05RR28 pKa = 11.84PRR30 pKa = 11.84MSGTWCPPVHH40 pKa = 6.17NVPGLEE46 pKa = 4.12RR47 pKa = 11.84NWYY50 pKa = 7.82EE51 pKa = 3.69SCLRR55 pKa = 11.84SHH57 pKa = 7.32AAFCGCGDD65 pKa = 4.38FVSHH69 pKa = 7.22LNNLANRR76 pKa = 11.84LGRR79 pKa = 11.84PPAPRR84 pKa = 11.84PPGAPQPPAVRR95 pKa = 11.84ALPALPAPEE104 pKa = 5.17DD105 pKa = 3.74RR106 pKa = 11.84FPNPPGWPGPGGGAAGAGAAAGRR129 pKa = 11.84DD130 pKa = 3.72GGDD133 pKa = 3.59GGDD136 pKa = 4.33AEE138 pKa = 5.32PGDD141 pKa = 4.43EE142 pKa = 5.76DD143 pKa = 5.86LDD145 pKa = 3.88ALFAADD151 pKa = 3.7EE152 pKa = 4.25
MM1 pKa = 7.45HH2 pKa = 7.29FSKK5 pKa = 10.89LSRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.51KK11 pKa = 10.58GKK13 pKa = 10.24VPLPPLPTAPLPPHH27 pKa = 7.05RR28 pKa = 11.84PRR30 pKa = 11.84MSGTWCPPVHH40 pKa = 6.17NVPGLEE46 pKa = 4.12RR47 pKa = 11.84NWYY50 pKa = 7.82EE51 pKa = 3.69SCLRR55 pKa = 11.84SHH57 pKa = 7.32AAFCGCGDD65 pKa = 4.38FVSHH69 pKa = 7.22LNNLANRR76 pKa = 11.84LGRR79 pKa = 11.84PPAPRR84 pKa = 11.84PPGAPQPPAVRR95 pKa = 11.84ALPALPAPEE104 pKa = 5.17DD105 pKa = 3.74RR106 pKa = 11.84FPNPPGWPGPGGGAAGAGAAAGRR129 pKa = 11.84DD130 pKa = 3.72GGDD133 pKa = 3.59GGDD136 pKa = 4.33AEE138 pKa = 5.32PGDD141 pKa = 4.43EE142 pKa = 5.76DD143 pKa = 5.86LDD145 pKa = 3.88ALFAADD151 pKa = 3.7EE152 pKa = 4.25
Molecular weight: 15.82 kDa
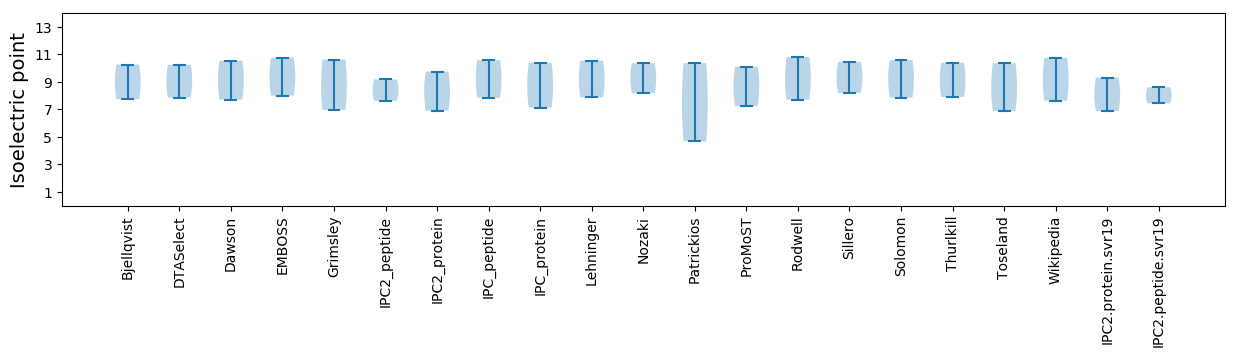
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9DUD0|ORF2_TTVV8 Uncharacterized ORF2 protein OS=Torque teno virus (isolate Chimpanzee/Japan/Pt-TTV6/2000) OX=687343 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.33AWPWRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84WRR11 pKa = 11.84WRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84WRR20 pKa = 11.84RR21 pKa = 11.84WRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84TWRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84PRR33 pKa = 11.84RR34 pKa = 11.84AVRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84VRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GRR46 pKa = 11.84GWRR49 pKa = 11.84RR50 pKa = 11.84LYY52 pKa = 10.01RR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 9.72RR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84GRR61 pKa = 11.84RR62 pKa = 11.84TYY64 pKa = 10.38KK65 pKa = 10.19KK66 pKa = 10.56KK67 pKa = 10.23PVLTQWQPTVRR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84CFIIGYY86 pKa = 8.07MPLIICGEE94 pKa = 4.01NLTSKK99 pKa = 10.65NYY101 pKa = 10.32ASHH104 pKa = 7.18ADD106 pKa = 3.85DD107 pKa = 4.11FVKK110 pKa = 10.66DD111 pKa = 3.94GPFGGGMTTMQFSLRR126 pKa = 11.84ILYY129 pKa = 10.41DD130 pKa = 3.14EE131 pKa = 4.94FLRR134 pKa = 11.84FLNIWTHH141 pKa = 5.67SNQDD145 pKa = 3.02LDD147 pKa = 3.6LARR150 pKa = 11.84YY151 pKa = 9.15HH152 pKa = 6.45GFKK155 pKa = 9.77LTLYY159 pKa = 9.52RR160 pKa = 11.84HH161 pKa = 5.57PTVDD165 pKa = 4.47FIVIIRR171 pKa = 11.84NSPPFEE177 pKa = 4.31DD178 pKa = 4.61TEE180 pKa = 4.32LTGPNTHH187 pKa = 7.26PGMLMLRR194 pKa = 11.84HH195 pKa = 6.04KK196 pKa = 10.36KK197 pKa = 9.84ILVPSLRR204 pKa = 11.84TRR206 pKa = 11.84PSRR209 pKa = 11.84RR210 pKa = 11.84HH211 pKa = 6.05KK212 pKa = 8.25ITVRR216 pKa = 11.84IGPPKK221 pKa = 10.27LFEE224 pKa = 5.05DD225 pKa = 3.23KK226 pKa = 10.07WYY228 pKa = 10.35SQTDD232 pKa = 3.08ICDD235 pKa = 3.6VILATVYY242 pKa = 9.35ATACDD247 pKa = 3.58LQYY250 pKa = 11.25PFGSPLTEE258 pKa = 3.94NYY260 pKa = 10.05CISFQVLGSAYY271 pKa = 10.89NNLISNTLNPNEE283 pKa = 4.08EE284 pKa = 4.01QQDD287 pKa = 3.57IKK289 pKa = 11.23SAIYY293 pKa = 10.74NNVNAYY299 pKa = 7.34LTRR302 pKa = 11.84ITEE305 pKa = 3.9SHH307 pKa = 5.93MANLISKK314 pKa = 9.52APPKK318 pKa = 10.15QVLKK322 pKa = 11.05SSDD325 pKa = 3.11GSLQTDD331 pKa = 3.34HH332 pKa = 7.29NDD334 pKa = 3.26TQFGGNPYY342 pKa = 8.64NTNQFTTTTVNKK354 pKa = 9.98IIQGAQNYY362 pKa = 7.11LTTIKK367 pKa = 9.04THH369 pKa = 5.96LQPNNSAINPNTQWHH384 pKa = 6.54LEE386 pKa = 4.03YY387 pKa = 10.58HH388 pKa = 6.12AGIYY392 pKa = 9.01SAPFLSAGRR401 pKa = 11.84LNPEE405 pKa = 3.63IKK407 pKa = 10.51GLYY410 pKa = 8.9TDD412 pKa = 3.47ITYY415 pKa = 11.14NPMMDD420 pKa = 3.55KK421 pKa = 10.51GTGNKK426 pKa = 8.6IWCDD430 pKa = 3.57SLTKK434 pKa = 10.59ADD436 pKa = 3.53MKK438 pKa = 9.87YY439 pKa = 8.58TEE441 pKa = 4.69GRR443 pKa = 11.84SKK445 pKa = 11.14YY446 pKa = 10.55LIEE449 pKa = 5.41NLPLWAAVWGYY460 pKa = 11.45LDD462 pKa = 4.26YY463 pKa = 10.3CTKK466 pKa = 10.36TSGDD470 pKa = 3.28AAFHH474 pKa = 5.35YY475 pKa = 10.1NYY477 pKa = 9.99RR478 pKa = 11.84VTLISPYY485 pKa = 9.05TSPMLFNPQDD495 pKa = 3.59PTKK498 pKa = 10.95GFVPYY503 pKa = 10.45SLNFGLGKK511 pKa = 9.64MPGGKK516 pKa = 9.99GYY518 pKa = 11.01VPLRR522 pKa = 11.84MRR524 pKa = 11.84ANWYY528 pKa = 9.35PYY530 pKa = 9.91FFHH533 pKa = 5.6QQKK536 pKa = 9.66VLEE539 pKa = 5.0AIGMSGPFTYY549 pKa = 10.41RR550 pKa = 11.84SDD552 pKa = 3.41EE553 pKa = 4.16KK554 pKa = 11.08KK555 pKa = 10.53AVLTSRR561 pKa = 11.84YY562 pKa = 9.32KK563 pKa = 10.85FKK565 pKa = 10.27FTWGGNPVSHH575 pKa = 5.32QVVRR579 pKa = 11.84NPCKK583 pKa = 9.65GTGGASASRR592 pKa = 11.84KK593 pKa = 7.95PRR595 pKa = 11.84SVQVTDD601 pKa = 3.86PKK603 pKa = 11.35YY604 pKa = 8.12NTPEE608 pKa = 4.03ITTHH612 pKa = 4.32TWDD615 pKa = 4.03IRR617 pKa = 11.84RR618 pKa = 11.84GWFGKK623 pKa = 10.35RR624 pKa = 11.84FIDD627 pKa = 5.16RR628 pKa = 11.84VQQQQASPEE637 pKa = 4.16LLADD641 pKa = 3.68PPKK644 pKa = 10.55RR645 pKa = 11.84PRR647 pKa = 11.84KK648 pKa = 8.7EE649 pKa = 3.73IKK651 pKa = 10.41GLTEE655 pKa = 4.36ADD657 pKa = 3.25QEE659 pKa = 4.3AEE661 pKa = 4.01KK662 pKa = 11.11DD663 pKa = 3.17SGLRR667 pKa = 11.84LRR669 pKa = 11.84QVQPWMSSQEE679 pKa = 4.2TQSEE683 pKa = 4.47QEE685 pKa = 4.33SAPEE689 pKa = 3.96EE690 pKa = 4.13QTVEE694 pKa = 3.91QQLRR698 pKa = 11.84NQLHH702 pKa = 4.85TQQLLGFQLRR712 pKa = 11.84SLMYY716 pKa = 9.84QVQQTHH722 pKa = 6.6RR723 pKa = 11.84NSFIHH728 pKa = 6.81PLLLPRR734 pKa = 11.84AA735 pKa = 4.06
MM1 pKa = 7.33AWPWRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84WRR11 pKa = 11.84WRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84WRR20 pKa = 11.84RR21 pKa = 11.84WRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84TWRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84PRR33 pKa = 11.84RR34 pKa = 11.84AVRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84VRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GRR46 pKa = 11.84GWRR49 pKa = 11.84RR50 pKa = 11.84LYY52 pKa = 10.01RR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 9.72RR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84GRR61 pKa = 11.84RR62 pKa = 11.84TYY64 pKa = 10.38KK65 pKa = 10.19KK66 pKa = 10.56KK67 pKa = 10.23PVLTQWQPTVRR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84CFIIGYY86 pKa = 8.07MPLIICGEE94 pKa = 4.01NLTSKK99 pKa = 10.65NYY101 pKa = 10.32ASHH104 pKa = 7.18ADD106 pKa = 3.85DD107 pKa = 4.11FVKK110 pKa = 10.66DD111 pKa = 3.94GPFGGGMTTMQFSLRR126 pKa = 11.84ILYY129 pKa = 10.41DD130 pKa = 3.14EE131 pKa = 4.94FLRR134 pKa = 11.84FLNIWTHH141 pKa = 5.67SNQDD145 pKa = 3.02LDD147 pKa = 3.6LARR150 pKa = 11.84YY151 pKa = 9.15HH152 pKa = 6.45GFKK155 pKa = 9.77LTLYY159 pKa = 9.52RR160 pKa = 11.84HH161 pKa = 5.57PTVDD165 pKa = 4.47FIVIIRR171 pKa = 11.84NSPPFEE177 pKa = 4.31DD178 pKa = 4.61TEE180 pKa = 4.32LTGPNTHH187 pKa = 7.26PGMLMLRR194 pKa = 11.84HH195 pKa = 6.04KK196 pKa = 10.36KK197 pKa = 9.84ILVPSLRR204 pKa = 11.84TRR206 pKa = 11.84PSRR209 pKa = 11.84RR210 pKa = 11.84HH211 pKa = 6.05KK212 pKa = 8.25ITVRR216 pKa = 11.84IGPPKK221 pKa = 10.27LFEE224 pKa = 5.05DD225 pKa = 3.23KK226 pKa = 10.07WYY228 pKa = 10.35SQTDD232 pKa = 3.08ICDD235 pKa = 3.6VILATVYY242 pKa = 9.35ATACDD247 pKa = 3.58LQYY250 pKa = 11.25PFGSPLTEE258 pKa = 3.94NYY260 pKa = 10.05CISFQVLGSAYY271 pKa = 10.89NNLISNTLNPNEE283 pKa = 4.08EE284 pKa = 4.01QQDD287 pKa = 3.57IKK289 pKa = 11.23SAIYY293 pKa = 10.74NNVNAYY299 pKa = 7.34LTRR302 pKa = 11.84ITEE305 pKa = 3.9SHH307 pKa = 5.93MANLISKK314 pKa = 9.52APPKK318 pKa = 10.15QVLKK322 pKa = 11.05SSDD325 pKa = 3.11GSLQTDD331 pKa = 3.34HH332 pKa = 7.29NDD334 pKa = 3.26TQFGGNPYY342 pKa = 8.64NTNQFTTTTVNKK354 pKa = 9.98IIQGAQNYY362 pKa = 7.11LTTIKK367 pKa = 9.04THH369 pKa = 5.96LQPNNSAINPNTQWHH384 pKa = 6.54LEE386 pKa = 4.03YY387 pKa = 10.58HH388 pKa = 6.12AGIYY392 pKa = 9.01SAPFLSAGRR401 pKa = 11.84LNPEE405 pKa = 3.63IKK407 pKa = 10.51GLYY410 pKa = 8.9TDD412 pKa = 3.47ITYY415 pKa = 11.14NPMMDD420 pKa = 3.55KK421 pKa = 10.51GTGNKK426 pKa = 8.6IWCDD430 pKa = 3.57SLTKK434 pKa = 10.59ADD436 pKa = 3.53MKK438 pKa = 9.87YY439 pKa = 8.58TEE441 pKa = 4.69GRR443 pKa = 11.84SKK445 pKa = 11.14YY446 pKa = 10.55LIEE449 pKa = 5.41NLPLWAAVWGYY460 pKa = 11.45LDD462 pKa = 4.26YY463 pKa = 10.3CTKK466 pKa = 10.36TSGDD470 pKa = 3.28AAFHH474 pKa = 5.35YY475 pKa = 10.1NYY477 pKa = 9.99RR478 pKa = 11.84VTLISPYY485 pKa = 9.05TSPMLFNPQDD495 pKa = 3.59PTKK498 pKa = 10.95GFVPYY503 pKa = 10.45SLNFGLGKK511 pKa = 9.64MPGGKK516 pKa = 9.99GYY518 pKa = 11.01VPLRR522 pKa = 11.84MRR524 pKa = 11.84ANWYY528 pKa = 9.35PYY530 pKa = 9.91FFHH533 pKa = 5.6QQKK536 pKa = 9.66VLEE539 pKa = 5.0AIGMSGPFTYY549 pKa = 10.41RR550 pKa = 11.84SDD552 pKa = 3.41EE553 pKa = 4.16KK554 pKa = 11.08KK555 pKa = 10.53AVLTSRR561 pKa = 11.84YY562 pKa = 9.32KK563 pKa = 10.85FKK565 pKa = 10.27FTWGGNPVSHH575 pKa = 5.32QVVRR579 pKa = 11.84NPCKK583 pKa = 9.65GTGGASASRR592 pKa = 11.84KK593 pKa = 7.95PRR595 pKa = 11.84SVQVTDD601 pKa = 3.86PKK603 pKa = 11.35YY604 pKa = 8.12NTPEE608 pKa = 4.03ITTHH612 pKa = 4.32TWDD615 pKa = 4.03IRR617 pKa = 11.84RR618 pKa = 11.84GWFGKK623 pKa = 10.35RR624 pKa = 11.84FIDD627 pKa = 5.16RR628 pKa = 11.84VQQQQASPEE637 pKa = 4.16LLADD641 pKa = 3.68PPKK644 pKa = 10.55RR645 pKa = 11.84PRR647 pKa = 11.84KK648 pKa = 8.7EE649 pKa = 3.73IKK651 pKa = 10.41GLTEE655 pKa = 4.36ADD657 pKa = 3.25QEE659 pKa = 4.3AEE661 pKa = 4.01KK662 pKa = 11.11DD663 pKa = 3.17SGLRR667 pKa = 11.84LRR669 pKa = 11.84QVQPWMSSQEE679 pKa = 4.2TQSEE683 pKa = 4.47QEE685 pKa = 4.33SAPEE689 pKa = 3.96EE690 pKa = 4.13QTVEE694 pKa = 3.91QQLRR698 pKa = 11.84NQLHH702 pKa = 4.85TQQLLGFQLRR712 pKa = 11.84SLMYY716 pKa = 9.84QVQQTHH722 pKa = 6.6RR723 pKa = 11.84NSFIHH728 pKa = 6.81PLLLPRR734 pKa = 11.84AA735 pKa = 4.06
Molecular weight: 86.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
887 |
152 |
735 |
443.5 |
50.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.088 ± 3.163 | 1.353 ± 0.572 |
4.397 ± 0.682 | 3.72 ± 0.102 |
3.72 ± 0.193 | 7.328 ± 2.608 |
2.706 ± 0.261 | 4.059 ± 1.816 |
4.961 ± 1.042 | 8.23 ± 0.144 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.029 ± 0.319 | 5.073 ± 0.504 |
8.794 ± 4.307 | 4.848 ± 1.874 |
9.921 ± 0.906 | 5.411 ± 0.655 |
6.652 ± 2.387 | 3.946 ± 0.294 |
2.593 ± 0.277 | 4.171 ± 1.572 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
