
Adonis mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Alphacarmovirus
Average proteome isoelectric point is 8.07
Get precalculated fractions of proteins
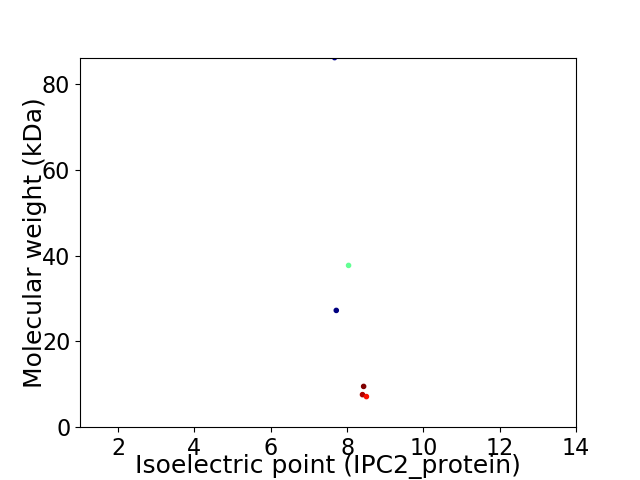
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
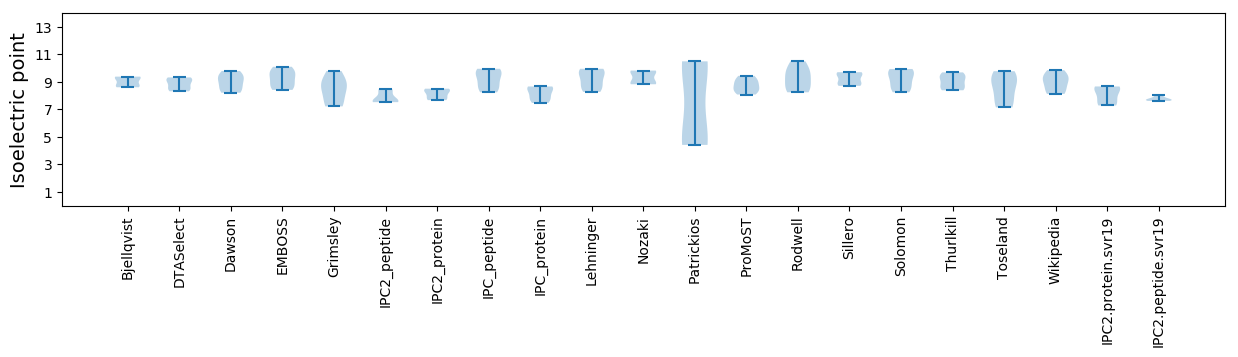
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J1E0V5|A0A1J1E0V5_9TOMB p7K OS=Adonis mosaic virus OX=1883104 GN=ORF5 PE=4 SV=1
MM1 pKa = 7.6GFLKK5 pKa = 10.47FATEE9 pKa = 4.0LAVGSTLMGGLVSVGIAGLTLRR31 pKa = 11.84ATIGVVEE38 pKa = 4.8FEE40 pKa = 3.92RR41 pKa = 11.84RR42 pKa = 11.84IIVGTCKK49 pKa = 10.28YY50 pKa = 9.36VASRR54 pKa = 11.84GLSLPSHH61 pKa = 7.0PEE63 pKa = 3.56YY64 pKa = 10.4PKK66 pKa = 11.19NSDD69 pKa = 3.25QLFLEE74 pKa = 4.77EE75 pKa = 4.29EE76 pKa = 4.43SEE78 pKa = 4.51SLSEE82 pKa = 4.13SQQVVEE88 pKa = 5.47DD89 pKa = 3.8FTEE92 pKa = 4.13LVEE95 pKa = 4.88EE96 pKa = 4.33KK97 pKa = 11.26GEE99 pKa = 4.09DD100 pKa = 3.55KK101 pKa = 10.73EE102 pKa = 4.36GKK104 pKa = 9.76EE105 pKa = 4.01VVISQKK111 pKa = 10.52RR112 pKa = 11.84VINRR116 pKa = 11.84HH117 pKa = 5.32SKK119 pKa = 10.44SKK121 pKa = 9.88FIRR124 pKa = 11.84YY125 pKa = 8.6IAIEE129 pKa = 4.22AKK131 pKa = 10.15NHH133 pKa = 6.75FGGDD137 pKa = 3.58LCASKK142 pKa = 11.13ANYY145 pKa = 10.25LSVSKK150 pKa = 10.47FLTGKK155 pKa = 9.81CKK157 pKa = 9.96EE158 pKa = 4.22RR159 pKa = 11.84QVVPAHH165 pKa = 5.77TRR167 pKa = 11.84VCVASAMALVFTPDD181 pKa = 3.03MAEE184 pKa = 3.72VQMRR188 pKa = 11.84VALNSHH194 pKa = 5.54EE195 pKa = 4.13AYY197 pKa = 9.7RR198 pKa = 11.84RR199 pKa = 11.84RR200 pKa = 11.84VAMEE204 pKa = 3.79AAGSMSSWGMRR215 pKa = 11.84LIQNPLTNSAWEE227 pKa = 4.08EE228 pKa = 3.26AWYY231 pKa = 10.33VLNGRR236 pKa = 11.84SSRR239 pKa = 11.84SAVTFTKK246 pKa = 10.64GGLFYY251 pKa = 11.38LEE253 pKa = 4.35GVEE256 pKa = 4.01TAIRR260 pKa = 11.84RR261 pKa = 11.84GSHH264 pKa = 6.03PSMIEE269 pKa = 3.93VEE271 pKa = 4.66LKK273 pKa = 10.74APPKK277 pKa = 10.03LRR279 pKa = 11.84KK280 pKa = 9.73LYY282 pKa = 9.57VQNSLSSGFDD292 pKa = 3.25YY293 pKa = 10.33RR294 pKa = 11.84VHH296 pKa = 5.42NHH298 pKa = 5.91SYY300 pKa = 9.54QNLRR304 pKa = 11.84RR305 pKa = 11.84GLLEE309 pKa = 3.47RR310 pKa = 11.84VFYY313 pKa = 11.26VEE315 pKa = 5.25SKK317 pKa = 10.77DD318 pKa = 3.93KK319 pKa = 11.08EE320 pKa = 4.32LVGCPRR326 pKa = 11.84PKK328 pKa = 10.2PGAFKK333 pKa = 10.6EE334 pKa = 3.83LGYY337 pKa = 10.43LRR339 pKa = 11.84RR340 pKa = 11.84RR341 pKa = 11.84FHH343 pKa = 7.32RR344 pKa = 11.84ICGNHH349 pKa = 4.54TRR351 pKa = 11.84ISAEE355 pKa = 3.95EE356 pKa = 4.05FVDD359 pKa = 4.84CYY361 pKa = 10.52QGRR364 pKa = 11.84KK365 pKa = 6.25RR366 pKa = 11.84TIYY369 pKa = 10.36EE370 pKa = 3.89NARR373 pKa = 11.84LSLDD377 pKa = 3.65DD378 pKa = 4.24KK379 pKa = 11.47ALEE382 pKa = 4.63RR383 pKa = 11.84KK384 pKa = 10.31DD385 pKa = 4.56GDD387 pKa = 3.54LKK389 pKa = 10.58TFIKK393 pKa = 10.68AEE395 pKa = 3.98KK396 pKa = 10.05FNISVKK402 pKa = 9.93PDD404 pKa = 3.51PAPRR408 pKa = 11.84VIQPRR413 pKa = 11.84SPRR416 pKa = 11.84FNLEE420 pKa = 3.47LGRR423 pKa = 11.84YY424 pKa = 7.93LKK426 pKa = 10.67KK427 pKa = 10.66FEE429 pKa = 4.61HH430 pKa = 6.28NAYY433 pKa = 9.74KK434 pKa = 10.49AIDD437 pKa = 4.56KK438 pKa = 10.37IWGAPTIMKK447 pKa = 10.17GYY449 pKa = 8.22NTEE452 pKa = 3.9EE453 pKa = 4.12VAQHH457 pKa = 6.81LKK459 pKa = 10.45HH460 pKa = 6.61AWDD463 pKa = 3.5QFQIPVAVGFDD474 pKa = 3.25MSRR477 pKa = 11.84FDD479 pKa = 3.32QHH481 pKa = 7.94VSVPALEE488 pKa = 4.93FEE490 pKa = 4.51HH491 pKa = 7.0SCYY494 pKa = 10.02TSCFEE499 pKa = 4.98GDD501 pKa = 3.18QHH503 pKa = 7.76LADD506 pKa = 4.48LLKK509 pKa = 10.13MQLKK513 pKa = 8.85NHH515 pKa = 5.75GVGFASNGMIRR526 pKa = 11.84YY527 pKa = 6.39TKK529 pKa = 9.82EE530 pKa = 3.57GCRR533 pKa = 11.84MSGDD537 pKa = 3.52MNTALGNCLLACLITKK553 pKa = 10.13HH554 pKa = 4.93ITRR557 pKa = 11.84GVRR560 pKa = 11.84GKK562 pKa = 10.71LINNGDD568 pKa = 3.87DD569 pKa = 3.53CVFICEE575 pKa = 4.19RR576 pKa = 11.84SDD578 pKa = 3.32AGSVVSDD585 pKa = 4.49LTTGWASFGFNCIAEE600 pKa = 4.47TPVEE604 pKa = 4.24VFEE607 pKa = 5.16EE608 pKa = 4.04IRR610 pKa = 11.84FCQMAPVFDD619 pKa = 4.13GEE621 pKa = 4.46VWVMVRR627 pKa = 11.84DD628 pKa = 4.2PKK630 pKa = 11.42VSMSKK635 pKa = 10.19DD636 pKa = 3.13AHH638 pKa = 7.1SLVHH642 pKa = 6.29WNNEE646 pKa = 3.59RR647 pKa = 11.84NALQWFKK654 pKa = 11.37AVGQCGMKK662 pKa = 9.78IAGRR666 pKa = 11.84VPVVQEE672 pKa = 3.95YY673 pKa = 7.1YY674 pKa = 10.31QKK676 pKa = 10.92YY677 pKa = 9.14IEE679 pKa = 4.5TGGDD683 pKa = 3.3VKK685 pKa = 10.43TSGSLSEE692 pKa = 4.2QVGGTGMFRR701 pKa = 11.84MAANSTRR708 pKa = 11.84GYY710 pKa = 10.73GAISQEE716 pKa = 3.76ARR718 pKa = 11.84YY719 pKa = 10.25SFYY722 pKa = 10.85LAFGIQPDD730 pKa = 3.67EE731 pKa = 4.37QIALEE736 pKa = 4.01HH737 pKa = 7.12EE738 pKa = 4.1IRR740 pKa = 11.84KK741 pKa = 5.64TTIHH745 pKa = 6.23ATLGPQCEE753 pKa = 4.56SADD756 pKa = 3.9SIQWLINPNLTT767 pKa = 3.6
MM1 pKa = 7.6GFLKK5 pKa = 10.47FATEE9 pKa = 4.0LAVGSTLMGGLVSVGIAGLTLRR31 pKa = 11.84ATIGVVEE38 pKa = 4.8FEE40 pKa = 3.92RR41 pKa = 11.84RR42 pKa = 11.84IIVGTCKK49 pKa = 10.28YY50 pKa = 9.36VASRR54 pKa = 11.84GLSLPSHH61 pKa = 7.0PEE63 pKa = 3.56YY64 pKa = 10.4PKK66 pKa = 11.19NSDD69 pKa = 3.25QLFLEE74 pKa = 4.77EE75 pKa = 4.29EE76 pKa = 4.43SEE78 pKa = 4.51SLSEE82 pKa = 4.13SQQVVEE88 pKa = 5.47DD89 pKa = 3.8FTEE92 pKa = 4.13LVEE95 pKa = 4.88EE96 pKa = 4.33KK97 pKa = 11.26GEE99 pKa = 4.09DD100 pKa = 3.55KK101 pKa = 10.73EE102 pKa = 4.36GKK104 pKa = 9.76EE105 pKa = 4.01VVISQKK111 pKa = 10.52RR112 pKa = 11.84VINRR116 pKa = 11.84HH117 pKa = 5.32SKK119 pKa = 10.44SKK121 pKa = 9.88FIRR124 pKa = 11.84YY125 pKa = 8.6IAIEE129 pKa = 4.22AKK131 pKa = 10.15NHH133 pKa = 6.75FGGDD137 pKa = 3.58LCASKK142 pKa = 11.13ANYY145 pKa = 10.25LSVSKK150 pKa = 10.47FLTGKK155 pKa = 9.81CKK157 pKa = 9.96EE158 pKa = 4.22RR159 pKa = 11.84QVVPAHH165 pKa = 5.77TRR167 pKa = 11.84VCVASAMALVFTPDD181 pKa = 3.03MAEE184 pKa = 3.72VQMRR188 pKa = 11.84VALNSHH194 pKa = 5.54EE195 pKa = 4.13AYY197 pKa = 9.7RR198 pKa = 11.84RR199 pKa = 11.84RR200 pKa = 11.84VAMEE204 pKa = 3.79AAGSMSSWGMRR215 pKa = 11.84LIQNPLTNSAWEE227 pKa = 4.08EE228 pKa = 3.26AWYY231 pKa = 10.33VLNGRR236 pKa = 11.84SSRR239 pKa = 11.84SAVTFTKK246 pKa = 10.64GGLFYY251 pKa = 11.38LEE253 pKa = 4.35GVEE256 pKa = 4.01TAIRR260 pKa = 11.84RR261 pKa = 11.84GSHH264 pKa = 6.03PSMIEE269 pKa = 3.93VEE271 pKa = 4.66LKK273 pKa = 10.74APPKK277 pKa = 10.03LRR279 pKa = 11.84KK280 pKa = 9.73LYY282 pKa = 9.57VQNSLSSGFDD292 pKa = 3.25YY293 pKa = 10.33RR294 pKa = 11.84VHH296 pKa = 5.42NHH298 pKa = 5.91SYY300 pKa = 9.54QNLRR304 pKa = 11.84RR305 pKa = 11.84GLLEE309 pKa = 3.47RR310 pKa = 11.84VFYY313 pKa = 11.26VEE315 pKa = 5.25SKK317 pKa = 10.77DD318 pKa = 3.93KK319 pKa = 11.08EE320 pKa = 4.32LVGCPRR326 pKa = 11.84PKK328 pKa = 10.2PGAFKK333 pKa = 10.6EE334 pKa = 3.83LGYY337 pKa = 10.43LRR339 pKa = 11.84RR340 pKa = 11.84RR341 pKa = 11.84FHH343 pKa = 7.32RR344 pKa = 11.84ICGNHH349 pKa = 4.54TRR351 pKa = 11.84ISAEE355 pKa = 3.95EE356 pKa = 4.05FVDD359 pKa = 4.84CYY361 pKa = 10.52QGRR364 pKa = 11.84KK365 pKa = 6.25RR366 pKa = 11.84TIYY369 pKa = 10.36EE370 pKa = 3.89NARR373 pKa = 11.84LSLDD377 pKa = 3.65DD378 pKa = 4.24KK379 pKa = 11.47ALEE382 pKa = 4.63RR383 pKa = 11.84KK384 pKa = 10.31DD385 pKa = 4.56GDD387 pKa = 3.54LKK389 pKa = 10.58TFIKK393 pKa = 10.68AEE395 pKa = 3.98KK396 pKa = 10.05FNISVKK402 pKa = 9.93PDD404 pKa = 3.51PAPRR408 pKa = 11.84VIQPRR413 pKa = 11.84SPRR416 pKa = 11.84FNLEE420 pKa = 3.47LGRR423 pKa = 11.84YY424 pKa = 7.93LKK426 pKa = 10.67KK427 pKa = 10.66FEE429 pKa = 4.61HH430 pKa = 6.28NAYY433 pKa = 9.74KK434 pKa = 10.49AIDD437 pKa = 4.56KK438 pKa = 10.37IWGAPTIMKK447 pKa = 10.17GYY449 pKa = 8.22NTEE452 pKa = 3.9EE453 pKa = 4.12VAQHH457 pKa = 6.81LKK459 pKa = 10.45HH460 pKa = 6.61AWDD463 pKa = 3.5QFQIPVAVGFDD474 pKa = 3.25MSRR477 pKa = 11.84FDD479 pKa = 3.32QHH481 pKa = 7.94VSVPALEE488 pKa = 4.93FEE490 pKa = 4.51HH491 pKa = 7.0SCYY494 pKa = 10.02TSCFEE499 pKa = 4.98GDD501 pKa = 3.18QHH503 pKa = 7.76LADD506 pKa = 4.48LLKK509 pKa = 10.13MQLKK513 pKa = 8.85NHH515 pKa = 5.75GVGFASNGMIRR526 pKa = 11.84YY527 pKa = 6.39TKK529 pKa = 9.82EE530 pKa = 3.57GCRR533 pKa = 11.84MSGDD537 pKa = 3.52MNTALGNCLLACLITKK553 pKa = 10.13HH554 pKa = 4.93ITRR557 pKa = 11.84GVRR560 pKa = 11.84GKK562 pKa = 10.71LINNGDD568 pKa = 3.87DD569 pKa = 3.53CVFICEE575 pKa = 4.19RR576 pKa = 11.84SDD578 pKa = 3.32AGSVVSDD585 pKa = 4.49LTTGWASFGFNCIAEE600 pKa = 4.47TPVEE604 pKa = 4.24VFEE607 pKa = 5.16EE608 pKa = 4.04IRR610 pKa = 11.84FCQMAPVFDD619 pKa = 4.13GEE621 pKa = 4.46VWVMVRR627 pKa = 11.84DD628 pKa = 4.2PKK630 pKa = 11.42VSMSKK635 pKa = 10.19DD636 pKa = 3.13AHH638 pKa = 7.1SLVHH642 pKa = 6.29WNNEE646 pKa = 3.59RR647 pKa = 11.84NALQWFKK654 pKa = 11.37AVGQCGMKK662 pKa = 9.78IAGRR666 pKa = 11.84VPVVQEE672 pKa = 3.95YY673 pKa = 7.1YY674 pKa = 10.31QKK676 pKa = 10.92YY677 pKa = 9.14IEE679 pKa = 4.5TGGDD683 pKa = 3.3VKK685 pKa = 10.43TSGSLSEE692 pKa = 4.2QVGGTGMFRR701 pKa = 11.84MAANSTRR708 pKa = 11.84GYY710 pKa = 10.73GAISQEE716 pKa = 3.76ARR718 pKa = 11.84YY719 pKa = 10.25SFYY722 pKa = 10.85LAFGIQPDD730 pKa = 3.67EE731 pKa = 4.37QIALEE736 pKa = 4.01HH737 pKa = 7.12EE738 pKa = 4.1IRR740 pKa = 11.84KK741 pKa = 5.64TTIHH745 pKa = 6.23ATLGPQCEE753 pKa = 4.56SADD756 pKa = 3.9SIQWLINPNLTT767 pKa = 3.6
Molecular weight: 86.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J1DWM6|A0A1J1DWM6_9TOMB RNA-directed RNA polymerase OS=Adonis mosaic virus OX=1883104 GN=ORF1RT PE=4 SV=1
MM1 pKa = 7.74PSANQNLTVITGVIGLLWLMRR22 pKa = 11.84SRR24 pKa = 11.84SILHH28 pKa = 6.65SIFEE32 pKa = 4.26AAKK35 pKa = 9.71PPPLLTLSQIIVFSFCGLLLNCISRR60 pKa = 11.84AEE62 pKa = 4.03RR63 pKa = 11.84EE64 pKa = 4.14VVITNNHH71 pKa = 6.62DD72 pKa = 3.49SSKK75 pKa = 9.64QQHH78 pKa = 5.51ITVNTPSKK86 pKa = 10.95
MM1 pKa = 7.74PSANQNLTVITGVIGLLWLMRR22 pKa = 11.84SRR24 pKa = 11.84SILHH28 pKa = 6.65SIFEE32 pKa = 4.26AAKK35 pKa = 9.71PPPLLTLSQIIVFSFCGLLLNCISRR60 pKa = 11.84AEE62 pKa = 4.03RR63 pKa = 11.84EE64 pKa = 4.14VVITNNHH71 pKa = 6.62DD72 pKa = 3.49SSKK75 pKa = 9.64QQHH78 pKa = 5.51ITVNTPSKK86 pKa = 10.95
Molecular weight: 9.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1576 |
64 |
767 |
262.7 |
29.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.741 ± 0.561 | 1.904 ± 0.413 |
3.744 ± 0.699 | 6.98 ± 0.911 |
4.569 ± 0.375 | 7.424 ± 0.77 |
2.411 ± 0.502 | 4.569 ± 0.821 |
6.091 ± 0.315 | 7.868 ± 0.587 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.538 ± 0.246 | 4.251 ± 0.711 |
3.934 ± 0.568 | 3.49 ± 0.276 |
6.218 ± 0.426 | 8.249 ± 0.829 |
5.266 ± 0.724 | 8.566 ± 0.678 |
1.586 ± 0.312 | 2.538 ± 0.616 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
