
Haemophilus phage Aaphi23
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
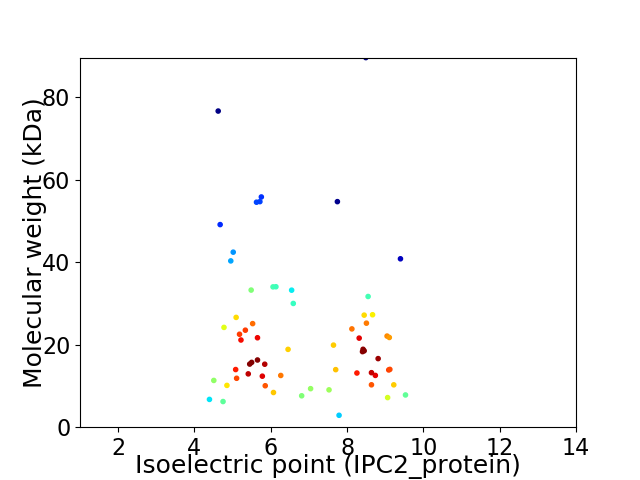
Virtual 2D-PAGE plot for 65 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
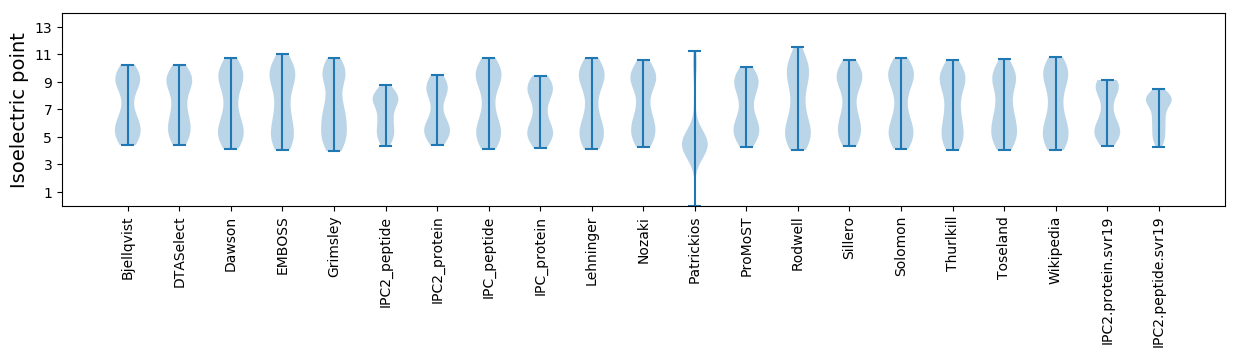
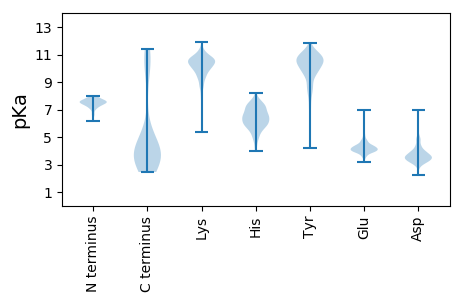
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q776W9|Q776W9_9CAUD Uncharacterized protein OS=Haemophilus phage Aaphi23 OX=230158 PE=4 SV=1
MM1 pKa = 7.67ANFIKK6 pKa = 10.63LSTINSVIFINMDD19 pKa = 4.35LIEE22 pKa = 4.63GMSRR26 pKa = 11.84DD27 pKa = 3.64DD28 pKa = 4.73SMTSLHH34 pKa = 6.75CSGDD38 pKa = 3.33ANSYY42 pKa = 9.87YY43 pKa = 10.42RR44 pKa = 11.84VTEE47 pKa = 4.0TPEE50 pKa = 4.36EE51 pKa = 3.81ILEE54 pKa = 4.22TMKK57 pKa = 10.9AAEE60 pKa = 4.04
MM1 pKa = 7.67ANFIKK6 pKa = 10.63LSTINSVIFINMDD19 pKa = 4.35LIEE22 pKa = 4.63GMSRR26 pKa = 11.84DD27 pKa = 3.64DD28 pKa = 4.73SMTSLHH34 pKa = 6.75CSGDD38 pKa = 3.33ANSYY42 pKa = 9.87YY43 pKa = 10.42RR44 pKa = 11.84VTEE47 pKa = 4.0TPEE50 pKa = 4.36EE51 pKa = 3.81ILEE54 pKa = 4.22TMKK57 pKa = 10.9AAEE60 pKa = 4.04
Molecular weight: 6.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q776X1|Q776X1_9CAUD Isoform of Q7Y5U8 Putative lytic protein Rz1 OS=Haemophilus phage Aaphi23 OX=230158 GN=terS PE=4 SV=1
MM1 pKa = 7.29SRR3 pKa = 11.84QIISLKK9 pKa = 9.91KK10 pKa = 10.04RR11 pKa = 11.84GEE13 pKa = 3.47IWHH16 pKa = 6.34YY17 pKa = 11.43SFTSPNGEE25 pKa = 3.62RR26 pKa = 11.84VRR28 pKa = 11.84RR29 pKa = 11.84SARR32 pKa = 11.84TSDD35 pKa = 3.34KK36 pKa = 11.32NQAQQLASKK45 pKa = 9.75EE46 pKa = 4.06YY47 pKa = 8.37NEE49 pKa = 4.29CWRR52 pKa = 11.84VFKK55 pKa = 11.01LGEE58 pKa = 4.02RR59 pKa = 11.84PNYY62 pKa = 8.46SWQEE66 pKa = 3.83AVVQWLDD73 pKa = 3.6EE74 pKa = 4.2KK75 pKa = 10.83PKK77 pKa = 10.56RR78 pKa = 11.84KK79 pKa = 8.93QDD81 pKa = 3.13RR82 pKa = 11.84NMLYY86 pKa = 11.0GLVWLDD92 pKa = 4.25KK93 pKa = 11.09YY94 pKa = 11.45LGDD97 pKa = 4.48KK98 pKa = 10.11KK99 pKa = 11.01LNEE102 pKa = 4.11IDD104 pKa = 3.12RR105 pKa = 11.84TLIKK109 pKa = 10.29FIQYY113 pKa = 8.83EE114 pKa = 4.13KK115 pKa = 10.78AKK117 pKa = 9.79EE118 pKa = 4.04GVKK121 pKa = 10.47ARR123 pKa = 11.84TINAVLQQIRR133 pKa = 11.84VILRR137 pKa = 11.84AAVEE141 pKa = 4.09WDD143 pKa = 3.28WLDD146 pKa = 3.26KK147 pKa = 11.23CPAIKK152 pKa = 10.16FLPEE156 pKa = 3.42PKK158 pKa = 9.93RR159 pKa = 11.84RR160 pKa = 11.84VRR162 pKa = 11.84WLTQHH167 pKa = 6.06EE168 pKa = 4.71EE169 pKa = 3.42IRR171 pKa = 11.84LIEE174 pKa = 4.33EE175 pKa = 4.41LPEE178 pKa = 3.93HH179 pKa = 6.92LKK181 pKa = 10.82PIVQFAILTGLRR193 pKa = 11.84MSNITQLKK201 pKa = 8.34WSQIDD206 pKa = 3.62LSKK209 pKa = 10.64RR210 pKa = 11.84QAWINSEE217 pKa = 3.89QSKK220 pKa = 8.83TGNSIGVPLNDD231 pKa = 3.74KK232 pKa = 10.4AIEE235 pKa = 4.33VIVSQFGKK243 pKa = 9.97HH244 pKa = 5.27KK245 pKa = 10.78EE246 pKa = 3.93NVFTYY251 pKa = 9.48KK252 pKa = 10.66GKK254 pKa = 9.54PVRR257 pKa = 11.84IANTKK262 pKa = 9.76AFRR265 pKa = 11.84AALQRR270 pKa = 11.84AGIKK274 pKa = 10.17DD275 pKa = 3.71FRR277 pKa = 11.84FHH279 pKa = 7.69DD280 pKa = 4.87LRR282 pKa = 11.84HH283 pKa = 5.01TWATRR288 pKa = 11.84HH289 pKa = 5.7IMSGTPLYY297 pKa = 10.62VLQEE301 pKa = 3.87LGGWSKK307 pKa = 11.67SDD309 pKa = 3.32TVRR312 pKa = 11.84KK313 pKa = 8.85YY314 pKa = 9.46AHH316 pKa = 6.89LSVEE320 pKa = 4.15HH321 pKa = 6.17LQNHH325 pKa = 6.43ANNVQLFATKK335 pKa = 10.39LAQTQRR341 pKa = 11.84GKK343 pKa = 10.57NLINNN348 pKa = 4.41
MM1 pKa = 7.29SRR3 pKa = 11.84QIISLKK9 pKa = 9.91KK10 pKa = 10.04RR11 pKa = 11.84GEE13 pKa = 3.47IWHH16 pKa = 6.34YY17 pKa = 11.43SFTSPNGEE25 pKa = 3.62RR26 pKa = 11.84VRR28 pKa = 11.84RR29 pKa = 11.84SARR32 pKa = 11.84TSDD35 pKa = 3.34KK36 pKa = 11.32NQAQQLASKK45 pKa = 9.75EE46 pKa = 4.06YY47 pKa = 8.37NEE49 pKa = 4.29CWRR52 pKa = 11.84VFKK55 pKa = 11.01LGEE58 pKa = 4.02RR59 pKa = 11.84PNYY62 pKa = 8.46SWQEE66 pKa = 3.83AVVQWLDD73 pKa = 3.6EE74 pKa = 4.2KK75 pKa = 10.83PKK77 pKa = 10.56RR78 pKa = 11.84KK79 pKa = 8.93QDD81 pKa = 3.13RR82 pKa = 11.84NMLYY86 pKa = 11.0GLVWLDD92 pKa = 4.25KK93 pKa = 11.09YY94 pKa = 11.45LGDD97 pKa = 4.48KK98 pKa = 10.11KK99 pKa = 11.01LNEE102 pKa = 4.11IDD104 pKa = 3.12RR105 pKa = 11.84TLIKK109 pKa = 10.29FIQYY113 pKa = 8.83EE114 pKa = 4.13KK115 pKa = 10.78AKK117 pKa = 9.79EE118 pKa = 4.04GVKK121 pKa = 10.47ARR123 pKa = 11.84TINAVLQQIRR133 pKa = 11.84VILRR137 pKa = 11.84AAVEE141 pKa = 4.09WDD143 pKa = 3.28WLDD146 pKa = 3.26KK147 pKa = 11.23CPAIKK152 pKa = 10.16FLPEE156 pKa = 3.42PKK158 pKa = 9.93RR159 pKa = 11.84RR160 pKa = 11.84VRR162 pKa = 11.84WLTQHH167 pKa = 6.06EE168 pKa = 4.71EE169 pKa = 3.42IRR171 pKa = 11.84LIEE174 pKa = 4.33EE175 pKa = 4.41LPEE178 pKa = 3.93HH179 pKa = 6.92LKK181 pKa = 10.82PIVQFAILTGLRR193 pKa = 11.84MSNITQLKK201 pKa = 8.34WSQIDD206 pKa = 3.62LSKK209 pKa = 10.64RR210 pKa = 11.84QAWINSEE217 pKa = 3.89QSKK220 pKa = 8.83TGNSIGVPLNDD231 pKa = 3.74KK232 pKa = 10.4AIEE235 pKa = 4.33VIVSQFGKK243 pKa = 9.97HH244 pKa = 5.27KK245 pKa = 10.78EE246 pKa = 3.93NVFTYY251 pKa = 9.48KK252 pKa = 10.66GKK254 pKa = 9.54PVRR257 pKa = 11.84IANTKK262 pKa = 9.76AFRR265 pKa = 11.84AALQRR270 pKa = 11.84AGIKK274 pKa = 10.17DD275 pKa = 3.71FRR277 pKa = 11.84FHH279 pKa = 7.69DD280 pKa = 4.87LRR282 pKa = 11.84HH283 pKa = 5.01TWATRR288 pKa = 11.84HH289 pKa = 5.7IMSGTPLYY297 pKa = 10.62VLQEE301 pKa = 3.87LGGWSKK307 pKa = 11.67SDD309 pKa = 3.32TVRR312 pKa = 11.84KK313 pKa = 8.85YY314 pKa = 9.46AHH316 pKa = 6.89LSVEE320 pKa = 4.15HH321 pKa = 6.17LQNHH325 pKa = 6.43ANNVQLFATKK335 pKa = 10.39LAQTQRR341 pKa = 11.84GKK343 pKa = 10.57NLINNN348 pKa = 4.41
Molecular weight: 40.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13554 |
27 |
800 |
208.5 |
23.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.175 ± 0.392 | 0.907 ± 0.112 |
5.799 ± 0.214 | 6.625 ± 0.322 |
4.25 ± 0.197 | 6.367 ± 0.269 |
1.601 ± 0.127 | 6.817 ± 0.201 |
7.769 ± 0.343 | 9.355 ± 0.29 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.206 ± 0.143 | 5.26 ± 0.194 |
2.973 ± 0.149 | 4.67 ± 0.201 |
4.663 ± 0.298 | 6.22 ± 0.215 |
5.829 ± 0.245 | 5.969 ± 0.181 |
1.409 ± 0.118 | 3.136 ± 0.18 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
