
Flavobacterium aquatile LMG 4008 = ATCC 11947
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; Flavobacterium aquatile
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
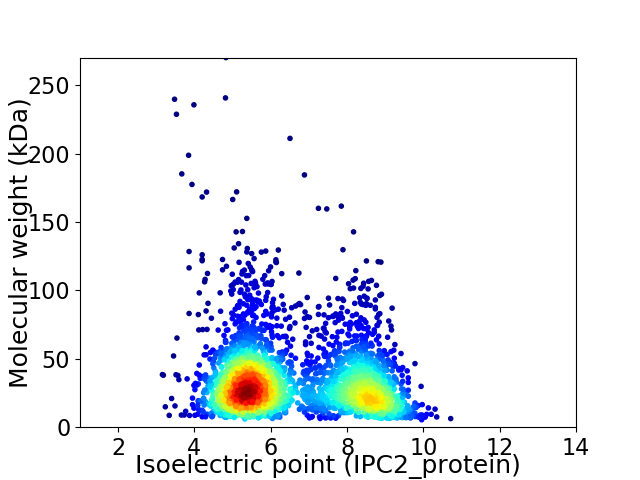
Virtual 2D-PAGE plot for 2995 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
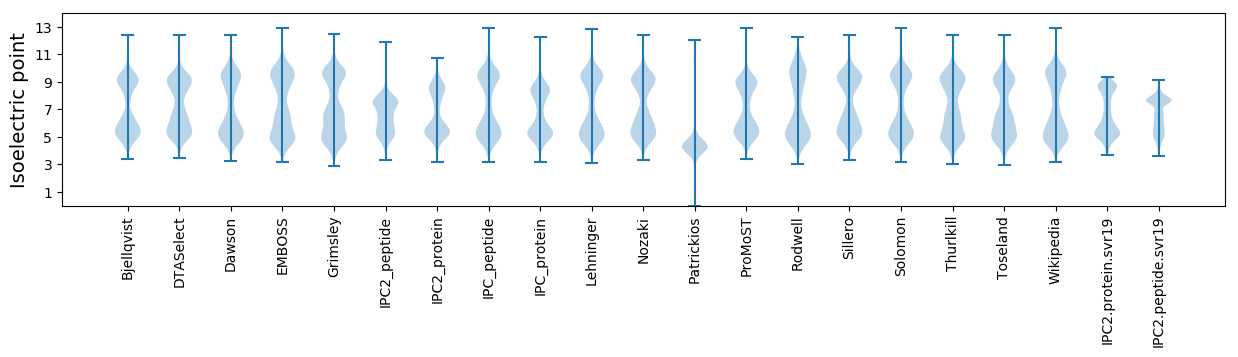
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A095TYF8|A0A095TYF8_9FLAO Mechanosensitive ion channel protein MscS OS=Flavobacterium aquatile LMG 4008 = ATCC 11947 OX=1453498 GN=LG45_14555 PE=4 SV=1
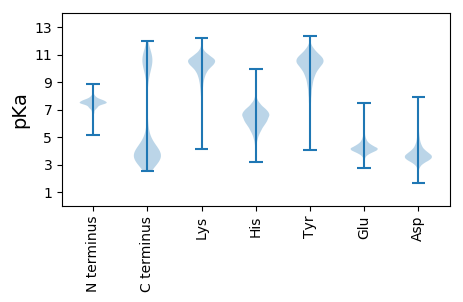
MM1 pKa = 7.11KK2 pKa = 10.27TNFKK6 pKa = 10.98NIGLVVLATLAFVSCSDD23 pKa = 3.93DD24 pKa = 3.81NPVEE28 pKa = 4.64GNQAPSAAEE37 pKa = 3.75FDD39 pKa = 4.07NVRR42 pKa = 11.84QTALDD47 pKa = 3.64NLTQNFTLVAEE58 pKa = 4.95DD59 pKa = 4.07GVTTFTSDD67 pKa = 2.86KK68 pKa = 10.67GVQFSINGNCLLKK81 pKa = 10.88NGNPVTGSVAIEE93 pKa = 3.88YY94 pKa = 10.78VEE96 pKa = 5.6LFDD99 pKa = 5.3AGNMLVTDD107 pKa = 4.28KK108 pKa = 9.0TTMGRR113 pKa = 11.84MPNGDD118 pKa = 3.23MTLLVSGGEE127 pKa = 4.17FYY129 pKa = 10.87INASQGGVDD138 pKa = 5.7LDD140 pKa = 3.83ITCPMQLLIPSTLTGGADD158 pKa = 2.96TGMTLWDD165 pKa = 3.54GTIDD169 pKa = 3.9EE170 pKa = 5.38DD171 pKa = 6.24GNLEE175 pKa = 4.12WDD177 pKa = 3.78EE178 pKa = 4.38QEE180 pKa = 4.66QNPAGQGGVFVEE192 pKa = 4.76GTGVNAPYY200 pKa = 10.65YY201 pKa = 11.01AFFDD205 pKa = 3.79SFGWTNVDD213 pKa = 3.64RR214 pKa = 11.84FYY216 pKa = 11.3SDD218 pKa = 3.14PRR220 pKa = 11.84PRR222 pKa = 11.84TMILAEE228 pKa = 4.55APNGYY233 pKa = 9.91DD234 pKa = 3.44FEE236 pKa = 5.39NSAVYY241 pKa = 9.76LHH243 pKa = 6.73YY244 pKa = 10.96DD245 pKa = 3.86GEE247 pKa = 4.74ASTLAKK253 pKa = 10.39LDD255 pKa = 3.76TYY257 pKa = 11.83NDD259 pKa = 3.28ATNQFSEE266 pKa = 5.04HH267 pKa = 6.39YY268 pKa = 8.61GQIPIGLEE276 pKa = 3.66CHH278 pKa = 6.32VIFVTEE284 pKa = 4.47EE285 pKa = 3.76DD286 pKa = 4.11GQWRR290 pKa = 11.84YY291 pKa = 10.08AIKK294 pKa = 10.47GVTIAADD301 pKa = 3.48DD302 pKa = 4.48VYY304 pKa = 11.16TFTLSEE310 pKa = 4.34TTVGSQAQLVAAINALPP327 pKa = 3.74
MM1 pKa = 7.11KK2 pKa = 10.27TNFKK6 pKa = 10.98NIGLVVLATLAFVSCSDD23 pKa = 3.93DD24 pKa = 3.81NPVEE28 pKa = 4.64GNQAPSAAEE37 pKa = 3.75FDD39 pKa = 4.07NVRR42 pKa = 11.84QTALDD47 pKa = 3.64NLTQNFTLVAEE58 pKa = 4.95DD59 pKa = 4.07GVTTFTSDD67 pKa = 2.86KK68 pKa = 10.67GVQFSINGNCLLKK81 pKa = 10.88NGNPVTGSVAIEE93 pKa = 3.88YY94 pKa = 10.78VEE96 pKa = 5.6LFDD99 pKa = 5.3AGNMLVTDD107 pKa = 4.28KK108 pKa = 9.0TTMGRR113 pKa = 11.84MPNGDD118 pKa = 3.23MTLLVSGGEE127 pKa = 4.17FYY129 pKa = 10.87INASQGGVDD138 pKa = 5.7LDD140 pKa = 3.83ITCPMQLLIPSTLTGGADD158 pKa = 2.96TGMTLWDD165 pKa = 3.54GTIDD169 pKa = 3.9EE170 pKa = 5.38DD171 pKa = 6.24GNLEE175 pKa = 4.12WDD177 pKa = 3.78EE178 pKa = 4.38QEE180 pKa = 4.66QNPAGQGGVFVEE192 pKa = 4.76GTGVNAPYY200 pKa = 10.65YY201 pKa = 11.01AFFDD205 pKa = 3.79SFGWTNVDD213 pKa = 3.64RR214 pKa = 11.84FYY216 pKa = 11.3SDD218 pKa = 3.14PRR220 pKa = 11.84PRR222 pKa = 11.84TMILAEE228 pKa = 4.55APNGYY233 pKa = 9.91DD234 pKa = 3.44FEE236 pKa = 5.39NSAVYY241 pKa = 9.76LHH243 pKa = 6.73YY244 pKa = 10.96DD245 pKa = 3.86GEE247 pKa = 4.74ASTLAKK253 pKa = 10.39LDD255 pKa = 3.76TYY257 pKa = 11.83NDD259 pKa = 3.28ATNQFSEE266 pKa = 5.04HH267 pKa = 6.39YY268 pKa = 8.61GQIPIGLEE276 pKa = 3.66CHH278 pKa = 6.32VIFVTEE284 pKa = 4.47EE285 pKa = 3.76DD286 pKa = 4.11GQWRR290 pKa = 11.84YY291 pKa = 10.08AIKK294 pKa = 10.47GVTIAADD301 pKa = 3.48DD302 pKa = 4.48VYY304 pKa = 11.16TFTLSEE310 pKa = 4.34TTVGSQAQLVAAINALPP327 pKa = 3.74
Molecular weight: 35.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A095UW20|A0A095UW20_9FLAO Uncharacterized protein OS=Flavobacterium aquatile LMG 4008 = ATCC 11947 OX=1453498 GN=LG45_15090 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
982436 |
46 |
2404 |
328.0 |
37.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.105 ± 0.047 | 0.779 ± 0.016 |
5.353 ± 0.031 | 6.469 ± 0.051 |
5.553 ± 0.039 | 6.056 ± 0.043 |
1.593 ± 0.021 | 8.373 ± 0.048 |
8.186 ± 0.065 | 9.077 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.19 ± 0.023 | 6.676 ± 0.056 |
3.27 ± 0.027 | 3.364 ± 0.025 |
3.028 ± 0.031 | 6.702 ± 0.042 |
5.969 ± 0.069 | 6.241 ± 0.038 |
0.972 ± 0.015 | 4.044 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
