
Gigaspora margarita mitovirus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
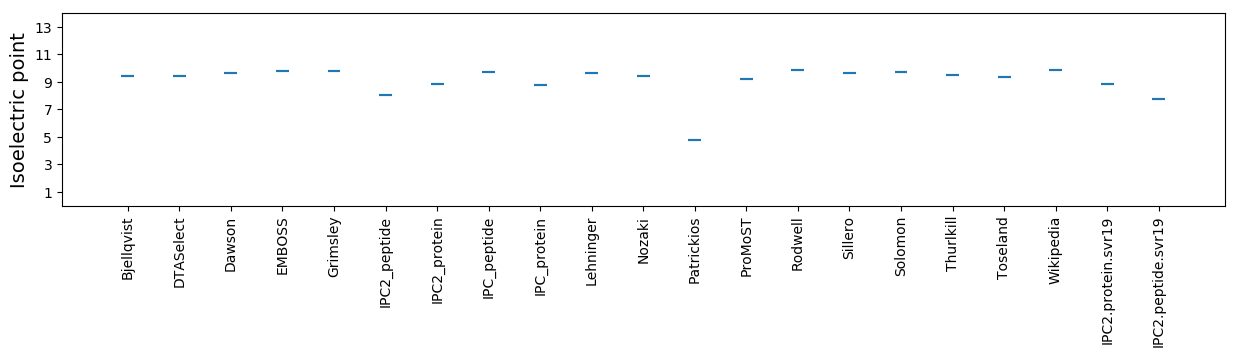
Average proteome isoelectric point is 8.82
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L0VZH5|A0A2L0VZH5_9VIRU RNA-dependent RNA polymerase OS=Gigaspora margarita mitovirus 3 OX=2082667 PE=4 SV=1
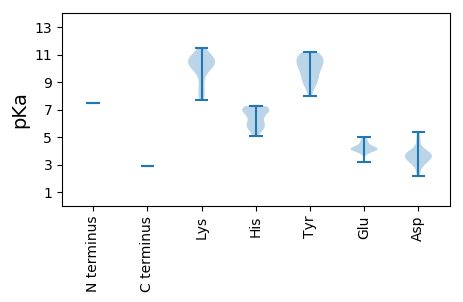
MM1 pKa = 7.48IRR3 pKa = 11.84LLINGPVPFGVTRR16 pKa = 11.84GAVNLSLINLQNALFSTSCVILRR39 pKa = 11.84KK40 pKa = 10.2YY41 pKa = 8.97EE42 pKa = 3.77VWTNLAKK49 pKa = 10.58RR50 pKa = 11.84HH51 pKa = 5.09GFAPAEE57 pKa = 4.23VIEE60 pKa = 4.32ASALGAYY67 pKa = 9.18VVVDD71 pKa = 3.86PMNKK75 pKa = 9.66SSLLALSPMAYY86 pKa = 9.59QKK88 pKa = 11.44LLGQTMSTEE97 pKa = 4.0NLSLILLLSPLDD109 pKa = 3.64EE110 pKa = 4.89LEE112 pKa = 4.23RR113 pKa = 11.84SAAEE117 pKa = 4.09ASPEE121 pKa = 4.05ATKK124 pKa = 10.8SPPLKK129 pKa = 10.69GFVNKK134 pKa = 10.18ISLDD138 pKa = 3.6LEE140 pKa = 4.09RR141 pKa = 11.84LKK143 pKa = 11.05EE144 pKa = 4.22RR145 pKa = 11.84IQTGRR150 pKa = 11.84LTKK153 pKa = 10.21GVRR156 pKa = 11.84WPSWSLGYY164 pKa = 9.67PFQGYY169 pKa = 9.29QGLGCRR175 pKa = 11.84LSDD178 pKa = 3.4GRR180 pKa = 11.84LILSRR185 pKa = 11.84PSEE188 pKa = 4.05FSAIFQRR195 pKa = 11.84WASRR199 pKa = 11.84LHH201 pKa = 5.58FWSGSSRR208 pKa = 11.84GRR210 pKa = 11.84RR211 pKa = 11.84FRR213 pKa = 11.84SSLWVVTNHH222 pKa = 5.85LQTVLKK228 pKa = 10.71HH229 pKa = 5.53SGPYY233 pKa = 10.23GLALYY238 pKa = 10.25LKK240 pKa = 9.8ASYY243 pKa = 10.66NIVLRR248 pKa = 11.84YY249 pKa = 9.6VAGQPFEE256 pKa = 4.58HH257 pKa = 7.22AFTQCGVPVQLQNGLPKK274 pKa = 10.4FLPRR278 pKa = 11.84EE279 pKa = 3.69WRR281 pKa = 11.84VAIRR285 pKa = 11.84SGSPRR290 pKa = 11.84LLRR293 pKa = 11.84IVLSLLYY300 pKa = 10.18SYY302 pKa = 11.16KK303 pKa = 10.53GFKK306 pKa = 8.19ATKK309 pKa = 8.28PKK311 pKa = 10.09PVAEE315 pKa = 3.9IFKK318 pKa = 10.25NISLAAHH325 pKa = 5.9QPDD328 pKa = 5.36DD329 pKa = 4.43IIAFSKK335 pKa = 10.84FCFEE339 pKa = 4.04FLKK342 pKa = 11.0NRR344 pKa = 11.84ISPISEE350 pKa = 4.05GEE352 pKa = 4.17LNPPSLPASLKK363 pKa = 10.54AGPNGPLSALAYY375 pKa = 10.34GADD378 pKa = 3.43AYY380 pKa = 10.9AWIQRR385 pKa = 11.84EE386 pKa = 4.13SAPYY390 pKa = 8.84QPSVPGPLEE399 pKa = 4.03WVQHH403 pKa = 5.8FGLDD407 pKa = 2.98KK408 pKa = 10.24WEE410 pKa = 4.29KK411 pKa = 10.44KK412 pKa = 9.31FRR414 pKa = 11.84AYY416 pKa = 10.95SDD418 pKa = 3.15WFLKK422 pKa = 10.62RR423 pKa = 11.84GFNHH427 pKa = 5.57QFNVTFPPPTRR438 pKa = 11.84KK439 pKa = 10.17GFGVLKK445 pKa = 10.54LKK447 pKa = 10.41IALGLNVPEE456 pKa = 4.78SKK458 pKa = 10.72SIALNPKK465 pKa = 8.92FAFHH469 pKa = 7.04RR470 pKa = 11.84RR471 pKa = 11.84TVSYY475 pKa = 10.0TPTRR479 pKa = 11.84VEE481 pKa = 4.24DD482 pKa = 4.27VEE484 pKa = 5.01ALQCLGLLLPVGRR497 pKa = 11.84VHH499 pKa = 6.76VLWEE503 pKa = 4.0AAGKK507 pKa = 7.82TRR509 pKa = 11.84VIAMMDD515 pKa = 3.94GLRR518 pKa = 11.84QALLRR523 pKa = 11.84PVHH526 pKa = 6.15LAMFRR531 pKa = 11.84RR532 pKa = 11.84IDD534 pKa = 3.83KK535 pKa = 10.84IFGFCSGLYY544 pKa = 9.18DD545 pKa = 3.08QSGAVRR551 pKa = 11.84SFAALNNRR559 pKa = 11.84EE560 pKa = 4.09VFSYY564 pKa = 10.47DD565 pKa = 2.51ISAATDD571 pKa = 3.86TIPYY575 pKa = 9.76QLYY578 pKa = 8.91FPLMEE583 pKa = 4.55FLLGKK588 pKa = 8.87TGAVLWNHH596 pKa = 5.95MVRR599 pKa = 11.84GHH601 pKa = 6.33PFIHH605 pKa = 6.93LEE607 pKa = 4.33GGDD610 pKa = 3.92EE611 pKa = 3.95PWAQRR616 pKa = 11.84LAGSSSYY623 pKa = 10.82YY624 pKa = 9.26YY625 pKa = 10.61EE626 pKa = 4.96RR627 pKa = 11.84GQPMGGYY634 pKa = 9.96SSFAALDD641 pKa = 3.84MLHH644 pKa = 6.74HH645 pKa = 7.09LIVQYY650 pKa = 10.96SAFEE654 pKa = 4.12AGYY657 pKa = 8.37DD658 pKa = 3.78TLNKK662 pKa = 9.88TFDD665 pKa = 3.26NYY667 pKa = 10.83RR668 pKa = 11.84ILGDD672 pKa = 3.74DD673 pKa = 3.63VVIGDD678 pKa = 3.48VRR680 pKa = 11.84VAKK683 pKa = 9.91TYY685 pKa = 11.11LNFMRR690 pKa = 11.84TWNIPISEE698 pKa = 4.25NKK700 pKa = 10.33SLVSSKK706 pKa = 11.01GVFQFLSEE714 pKa = 4.01VFRR717 pKa = 11.84GGEE720 pKa = 4.15CLSPLSFRR728 pKa = 11.84ADD730 pKa = 3.64YY731 pKa = 10.43QSKK734 pKa = 8.02TFLEE738 pKa = 4.41RR739 pKa = 11.84FSFAATAVSRR749 pKa = 11.84GWSSGLAMEE758 pKa = 5.02GFGRR762 pKa = 11.84LILSKK767 pKa = 10.89DD768 pKa = 3.27QFLSEE773 pKa = 3.22IHH775 pKa = 7.07RR776 pKa = 11.84YY777 pKa = 8.66RR778 pKa = 11.84NRR780 pKa = 11.84RR781 pKa = 11.84PSAFIYY787 pKa = 10.59SLLFALVTFAGEE799 pKa = 4.18AGWALGSWLHH809 pKa = 5.92NVVAHH814 pKa = 7.25DD815 pKa = 4.96PFPQLLSNKK824 pKa = 7.69EE825 pKa = 4.09LEE827 pKa = 4.48SVEE830 pKa = 4.22SKK832 pKa = 11.2NFVFTILRR840 pKa = 11.84QILFTEE846 pKa = 4.72AGRR849 pKa = 11.84IHH851 pKa = 6.98EE852 pKa = 4.19LQKK855 pKa = 11.11AEE857 pKa = 4.16SGVNQTRR864 pKa = 11.84QFLDD868 pKa = 3.71FGSIEE873 pKa = 3.91FHH875 pKa = 6.95NDD877 pKa = 2.19IEE879 pKa = 4.72NDD881 pKa = 3.43VDD883 pKa = 4.9SLWLLARR890 pKa = 11.84EE891 pKa = 4.43GKK893 pKa = 8.11TLLEE897 pKa = 4.55SADD900 pKa = 3.74VTPDD904 pKa = 2.7WVTSAFSYY912 pKa = 9.98LRR914 pKa = 11.84KK915 pKa = 9.97VSGIIPRR922 pKa = 11.84LSATGVVHH930 pKa = 6.72PLDD933 pKa = 3.88YY934 pKa = 10.98LVLMKK939 pKa = 8.87TTEE942 pKa = 3.97DD943 pKa = 3.29RR944 pKa = 11.84NNIHH948 pKa = 7.14KK949 pKa = 10.22VNQWSPTNHH958 pKa = 6.65DD959 pKa = 3.27AEE961 pKa = 4.62EE962 pKa = 4.07IFRR965 pKa = 11.84VRR967 pKa = 11.84MVDD970 pKa = 3.26TTFSLIKK977 pKa = 10.4SNPYY981 pKa = 8.01YY982 pKa = 10.8HH983 pKa = 7.13EE984 pKa = 4.31VLTGISSPYY993 pKa = 10.21GLALRR998 pKa = 11.84PVPEE1002 pKa = 4.27EE1003 pKa = 3.89GRR1005 pKa = 11.84PSGSSS1010 pKa = 2.91
MM1 pKa = 7.48IRR3 pKa = 11.84LLINGPVPFGVTRR16 pKa = 11.84GAVNLSLINLQNALFSTSCVILRR39 pKa = 11.84KK40 pKa = 10.2YY41 pKa = 8.97EE42 pKa = 3.77VWTNLAKK49 pKa = 10.58RR50 pKa = 11.84HH51 pKa = 5.09GFAPAEE57 pKa = 4.23VIEE60 pKa = 4.32ASALGAYY67 pKa = 9.18VVVDD71 pKa = 3.86PMNKK75 pKa = 9.66SSLLALSPMAYY86 pKa = 9.59QKK88 pKa = 11.44LLGQTMSTEE97 pKa = 4.0NLSLILLLSPLDD109 pKa = 3.64EE110 pKa = 4.89LEE112 pKa = 4.23RR113 pKa = 11.84SAAEE117 pKa = 4.09ASPEE121 pKa = 4.05ATKK124 pKa = 10.8SPPLKK129 pKa = 10.69GFVNKK134 pKa = 10.18ISLDD138 pKa = 3.6LEE140 pKa = 4.09RR141 pKa = 11.84LKK143 pKa = 11.05EE144 pKa = 4.22RR145 pKa = 11.84IQTGRR150 pKa = 11.84LTKK153 pKa = 10.21GVRR156 pKa = 11.84WPSWSLGYY164 pKa = 9.67PFQGYY169 pKa = 9.29QGLGCRR175 pKa = 11.84LSDD178 pKa = 3.4GRR180 pKa = 11.84LILSRR185 pKa = 11.84PSEE188 pKa = 4.05FSAIFQRR195 pKa = 11.84WASRR199 pKa = 11.84LHH201 pKa = 5.58FWSGSSRR208 pKa = 11.84GRR210 pKa = 11.84RR211 pKa = 11.84FRR213 pKa = 11.84SSLWVVTNHH222 pKa = 5.85LQTVLKK228 pKa = 10.71HH229 pKa = 5.53SGPYY233 pKa = 10.23GLALYY238 pKa = 10.25LKK240 pKa = 9.8ASYY243 pKa = 10.66NIVLRR248 pKa = 11.84YY249 pKa = 9.6VAGQPFEE256 pKa = 4.58HH257 pKa = 7.22AFTQCGVPVQLQNGLPKK274 pKa = 10.4FLPRR278 pKa = 11.84EE279 pKa = 3.69WRR281 pKa = 11.84VAIRR285 pKa = 11.84SGSPRR290 pKa = 11.84LLRR293 pKa = 11.84IVLSLLYY300 pKa = 10.18SYY302 pKa = 11.16KK303 pKa = 10.53GFKK306 pKa = 8.19ATKK309 pKa = 8.28PKK311 pKa = 10.09PVAEE315 pKa = 3.9IFKK318 pKa = 10.25NISLAAHH325 pKa = 5.9QPDD328 pKa = 5.36DD329 pKa = 4.43IIAFSKK335 pKa = 10.84FCFEE339 pKa = 4.04FLKK342 pKa = 11.0NRR344 pKa = 11.84ISPISEE350 pKa = 4.05GEE352 pKa = 4.17LNPPSLPASLKK363 pKa = 10.54AGPNGPLSALAYY375 pKa = 10.34GADD378 pKa = 3.43AYY380 pKa = 10.9AWIQRR385 pKa = 11.84EE386 pKa = 4.13SAPYY390 pKa = 8.84QPSVPGPLEE399 pKa = 4.03WVQHH403 pKa = 5.8FGLDD407 pKa = 2.98KK408 pKa = 10.24WEE410 pKa = 4.29KK411 pKa = 10.44KK412 pKa = 9.31FRR414 pKa = 11.84AYY416 pKa = 10.95SDD418 pKa = 3.15WFLKK422 pKa = 10.62RR423 pKa = 11.84GFNHH427 pKa = 5.57QFNVTFPPPTRR438 pKa = 11.84KK439 pKa = 10.17GFGVLKK445 pKa = 10.54LKK447 pKa = 10.41IALGLNVPEE456 pKa = 4.78SKK458 pKa = 10.72SIALNPKK465 pKa = 8.92FAFHH469 pKa = 7.04RR470 pKa = 11.84RR471 pKa = 11.84TVSYY475 pKa = 10.0TPTRR479 pKa = 11.84VEE481 pKa = 4.24DD482 pKa = 4.27VEE484 pKa = 5.01ALQCLGLLLPVGRR497 pKa = 11.84VHH499 pKa = 6.76VLWEE503 pKa = 4.0AAGKK507 pKa = 7.82TRR509 pKa = 11.84VIAMMDD515 pKa = 3.94GLRR518 pKa = 11.84QALLRR523 pKa = 11.84PVHH526 pKa = 6.15LAMFRR531 pKa = 11.84RR532 pKa = 11.84IDD534 pKa = 3.83KK535 pKa = 10.84IFGFCSGLYY544 pKa = 9.18DD545 pKa = 3.08QSGAVRR551 pKa = 11.84SFAALNNRR559 pKa = 11.84EE560 pKa = 4.09VFSYY564 pKa = 10.47DD565 pKa = 2.51ISAATDD571 pKa = 3.86TIPYY575 pKa = 9.76QLYY578 pKa = 8.91FPLMEE583 pKa = 4.55FLLGKK588 pKa = 8.87TGAVLWNHH596 pKa = 5.95MVRR599 pKa = 11.84GHH601 pKa = 6.33PFIHH605 pKa = 6.93LEE607 pKa = 4.33GGDD610 pKa = 3.92EE611 pKa = 3.95PWAQRR616 pKa = 11.84LAGSSSYY623 pKa = 10.82YY624 pKa = 9.26YY625 pKa = 10.61EE626 pKa = 4.96RR627 pKa = 11.84GQPMGGYY634 pKa = 9.96SSFAALDD641 pKa = 3.84MLHH644 pKa = 6.74HH645 pKa = 7.09LIVQYY650 pKa = 10.96SAFEE654 pKa = 4.12AGYY657 pKa = 8.37DD658 pKa = 3.78TLNKK662 pKa = 9.88TFDD665 pKa = 3.26NYY667 pKa = 10.83RR668 pKa = 11.84ILGDD672 pKa = 3.74DD673 pKa = 3.63VVIGDD678 pKa = 3.48VRR680 pKa = 11.84VAKK683 pKa = 9.91TYY685 pKa = 11.11LNFMRR690 pKa = 11.84TWNIPISEE698 pKa = 4.25NKK700 pKa = 10.33SLVSSKK706 pKa = 11.01GVFQFLSEE714 pKa = 4.01VFRR717 pKa = 11.84GGEE720 pKa = 4.15CLSPLSFRR728 pKa = 11.84ADD730 pKa = 3.64YY731 pKa = 10.43QSKK734 pKa = 8.02TFLEE738 pKa = 4.41RR739 pKa = 11.84FSFAATAVSRR749 pKa = 11.84GWSSGLAMEE758 pKa = 5.02GFGRR762 pKa = 11.84LILSKK767 pKa = 10.89DD768 pKa = 3.27QFLSEE773 pKa = 3.22IHH775 pKa = 7.07RR776 pKa = 11.84YY777 pKa = 8.66RR778 pKa = 11.84NRR780 pKa = 11.84RR781 pKa = 11.84PSAFIYY787 pKa = 10.59SLLFALVTFAGEE799 pKa = 4.18AGWALGSWLHH809 pKa = 5.92NVVAHH814 pKa = 7.25DD815 pKa = 4.96PFPQLLSNKK824 pKa = 7.69EE825 pKa = 4.09LEE827 pKa = 4.48SVEE830 pKa = 4.22SKK832 pKa = 11.2NFVFTILRR840 pKa = 11.84QILFTEE846 pKa = 4.72AGRR849 pKa = 11.84IHH851 pKa = 6.98EE852 pKa = 4.19LQKK855 pKa = 11.11AEE857 pKa = 4.16SGVNQTRR864 pKa = 11.84QFLDD868 pKa = 3.71FGSIEE873 pKa = 3.91FHH875 pKa = 6.95NDD877 pKa = 2.19IEE879 pKa = 4.72NDD881 pKa = 3.43VDD883 pKa = 4.9SLWLLARR890 pKa = 11.84EE891 pKa = 4.43GKK893 pKa = 8.11TLLEE897 pKa = 4.55SADD900 pKa = 3.74VTPDD904 pKa = 2.7WVTSAFSYY912 pKa = 9.98LRR914 pKa = 11.84KK915 pKa = 9.97VSGIIPRR922 pKa = 11.84LSATGVVHH930 pKa = 6.72PLDD933 pKa = 3.88YY934 pKa = 10.98LVLMKK939 pKa = 8.87TTEE942 pKa = 3.97DD943 pKa = 3.29RR944 pKa = 11.84NNIHH948 pKa = 7.14KK949 pKa = 10.22VNQWSPTNHH958 pKa = 6.65DD959 pKa = 3.27AEE961 pKa = 4.62EE962 pKa = 4.07IFRR965 pKa = 11.84VRR967 pKa = 11.84MVDD970 pKa = 3.26TTFSLIKK977 pKa = 10.4SNPYY981 pKa = 8.01YY982 pKa = 10.8HH983 pKa = 7.13EE984 pKa = 4.31VLTGISSPYY993 pKa = 10.21GLALRR998 pKa = 11.84PVPEE1002 pKa = 4.27EE1003 pKa = 3.89GRR1005 pKa = 11.84PSGSSS1010 pKa = 2.91
Molecular weight: 113.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L0VZH5|A0A2L0VZH5_9VIRU RNA-dependent RNA polymerase OS=Gigaspora margarita mitovirus 3 OX=2082667 PE=4 SV=1
MM1 pKa = 7.48IRR3 pKa = 11.84LLINGPVPFGVTRR16 pKa = 11.84GAVNLSLINLQNALFSTSCVILRR39 pKa = 11.84KK40 pKa = 10.2YY41 pKa = 8.97EE42 pKa = 3.77VWTNLAKK49 pKa = 10.58RR50 pKa = 11.84HH51 pKa = 5.09GFAPAEE57 pKa = 4.23VIEE60 pKa = 4.32ASALGAYY67 pKa = 9.18VVVDD71 pKa = 3.86PMNKK75 pKa = 9.66SSLLALSPMAYY86 pKa = 9.59QKK88 pKa = 11.44LLGQTMSTEE97 pKa = 4.0NLSLILLLSPLDD109 pKa = 3.64EE110 pKa = 4.89LEE112 pKa = 4.23RR113 pKa = 11.84SAAEE117 pKa = 4.09ASPEE121 pKa = 4.05ATKK124 pKa = 10.8SPPLKK129 pKa = 10.69GFVNKK134 pKa = 10.18ISLDD138 pKa = 3.6LEE140 pKa = 4.09RR141 pKa = 11.84LKK143 pKa = 11.05EE144 pKa = 4.22RR145 pKa = 11.84IQTGRR150 pKa = 11.84LTKK153 pKa = 10.21GVRR156 pKa = 11.84WPSWSLGYY164 pKa = 9.67PFQGYY169 pKa = 9.29QGLGCRR175 pKa = 11.84LSDD178 pKa = 3.4GRR180 pKa = 11.84LILSRR185 pKa = 11.84PSEE188 pKa = 4.05FSAIFQRR195 pKa = 11.84WASRR199 pKa = 11.84LHH201 pKa = 5.58FWSGSSRR208 pKa = 11.84GRR210 pKa = 11.84RR211 pKa = 11.84FRR213 pKa = 11.84SSLWVVTNHH222 pKa = 5.85LQTVLKK228 pKa = 10.71HH229 pKa = 5.53SGPYY233 pKa = 10.23GLALYY238 pKa = 10.25LKK240 pKa = 9.8ASYY243 pKa = 10.66NIVLRR248 pKa = 11.84YY249 pKa = 9.6VAGQPFEE256 pKa = 4.58HH257 pKa = 7.22AFTQCGVPVQLQNGLPKK274 pKa = 10.4FLPRR278 pKa = 11.84EE279 pKa = 3.69WRR281 pKa = 11.84VAIRR285 pKa = 11.84SGSPRR290 pKa = 11.84LLRR293 pKa = 11.84IVLSLLYY300 pKa = 10.18SYY302 pKa = 11.16KK303 pKa = 10.53GFKK306 pKa = 8.19ATKK309 pKa = 8.28PKK311 pKa = 10.09PVAEE315 pKa = 3.9IFKK318 pKa = 10.25NISLAAHH325 pKa = 5.9QPDD328 pKa = 5.36DD329 pKa = 4.43IIAFSKK335 pKa = 10.84FCFEE339 pKa = 4.04FLKK342 pKa = 11.0NRR344 pKa = 11.84ISPISEE350 pKa = 4.05GEE352 pKa = 4.17LNPPSLPASLKK363 pKa = 10.54AGPNGPLSALAYY375 pKa = 10.34GADD378 pKa = 3.43AYY380 pKa = 10.9AWIQRR385 pKa = 11.84EE386 pKa = 4.13SAPYY390 pKa = 8.84QPSVPGPLEE399 pKa = 4.03WVQHH403 pKa = 5.8FGLDD407 pKa = 2.98KK408 pKa = 10.24WEE410 pKa = 4.29KK411 pKa = 10.44KK412 pKa = 9.31FRR414 pKa = 11.84AYY416 pKa = 10.95SDD418 pKa = 3.15WFLKK422 pKa = 10.62RR423 pKa = 11.84GFNHH427 pKa = 5.57QFNVTFPPPTRR438 pKa = 11.84KK439 pKa = 10.17GFGVLKK445 pKa = 10.54LKK447 pKa = 10.41IALGLNVPEE456 pKa = 4.78SKK458 pKa = 10.72SIALNPKK465 pKa = 8.92FAFHH469 pKa = 7.04RR470 pKa = 11.84RR471 pKa = 11.84TVSYY475 pKa = 10.0TPTRR479 pKa = 11.84VEE481 pKa = 4.24DD482 pKa = 4.27VEE484 pKa = 5.01ALQCLGLLLPVGRR497 pKa = 11.84VHH499 pKa = 6.76VLWEE503 pKa = 4.0AAGKK507 pKa = 7.82TRR509 pKa = 11.84VIAMMDD515 pKa = 3.94GLRR518 pKa = 11.84QALLRR523 pKa = 11.84PVHH526 pKa = 6.15LAMFRR531 pKa = 11.84RR532 pKa = 11.84IDD534 pKa = 3.83KK535 pKa = 10.84IFGFCSGLYY544 pKa = 9.18DD545 pKa = 3.08QSGAVRR551 pKa = 11.84SFAALNNRR559 pKa = 11.84EE560 pKa = 4.09VFSYY564 pKa = 10.47DD565 pKa = 2.51ISAATDD571 pKa = 3.86TIPYY575 pKa = 9.76QLYY578 pKa = 8.91FPLMEE583 pKa = 4.55FLLGKK588 pKa = 8.87TGAVLWNHH596 pKa = 5.95MVRR599 pKa = 11.84GHH601 pKa = 6.33PFIHH605 pKa = 6.93LEE607 pKa = 4.33GGDD610 pKa = 3.92EE611 pKa = 3.95PWAQRR616 pKa = 11.84LAGSSSYY623 pKa = 10.82YY624 pKa = 9.26YY625 pKa = 10.61EE626 pKa = 4.96RR627 pKa = 11.84GQPMGGYY634 pKa = 9.96SSFAALDD641 pKa = 3.84MLHH644 pKa = 6.74HH645 pKa = 7.09LIVQYY650 pKa = 10.96SAFEE654 pKa = 4.12AGYY657 pKa = 8.37DD658 pKa = 3.78TLNKK662 pKa = 9.88TFDD665 pKa = 3.26NYY667 pKa = 10.83RR668 pKa = 11.84ILGDD672 pKa = 3.74DD673 pKa = 3.63VVIGDD678 pKa = 3.48VRR680 pKa = 11.84VAKK683 pKa = 9.91TYY685 pKa = 11.11LNFMRR690 pKa = 11.84TWNIPISEE698 pKa = 4.25NKK700 pKa = 10.33SLVSSKK706 pKa = 11.01GVFQFLSEE714 pKa = 4.01VFRR717 pKa = 11.84GGEE720 pKa = 4.15CLSPLSFRR728 pKa = 11.84ADD730 pKa = 3.64YY731 pKa = 10.43QSKK734 pKa = 8.02TFLEE738 pKa = 4.41RR739 pKa = 11.84FSFAATAVSRR749 pKa = 11.84GWSSGLAMEE758 pKa = 5.02GFGRR762 pKa = 11.84LILSKK767 pKa = 10.89DD768 pKa = 3.27QFLSEE773 pKa = 3.22IHH775 pKa = 7.07RR776 pKa = 11.84YY777 pKa = 8.66RR778 pKa = 11.84NRR780 pKa = 11.84RR781 pKa = 11.84PSAFIYY787 pKa = 10.59SLLFALVTFAGEE799 pKa = 4.18AGWALGSWLHH809 pKa = 5.92NVVAHH814 pKa = 7.25DD815 pKa = 4.96PFPQLLSNKK824 pKa = 7.69EE825 pKa = 4.09LEE827 pKa = 4.48SVEE830 pKa = 4.22SKK832 pKa = 11.2NFVFTILRR840 pKa = 11.84QILFTEE846 pKa = 4.72AGRR849 pKa = 11.84IHH851 pKa = 6.98EE852 pKa = 4.19LQKK855 pKa = 11.11AEE857 pKa = 4.16SGVNQTRR864 pKa = 11.84QFLDD868 pKa = 3.71FGSIEE873 pKa = 3.91FHH875 pKa = 6.95NDD877 pKa = 2.19IEE879 pKa = 4.72NDD881 pKa = 3.43VDD883 pKa = 4.9SLWLLARR890 pKa = 11.84EE891 pKa = 4.43GKK893 pKa = 8.11TLLEE897 pKa = 4.55SADD900 pKa = 3.74VTPDD904 pKa = 2.7WVTSAFSYY912 pKa = 9.98LRR914 pKa = 11.84KK915 pKa = 9.97VSGIIPRR922 pKa = 11.84LSATGVVHH930 pKa = 6.72PLDD933 pKa = 3.88YY934 pKa = 10.98LVLMKK939 pKa = 8.87TTEE942 pKa = 3.97DD943 pKa = 3.29RR944 pKa = 11.84NNIHH948 pKa = 7.14KK949 pKa = 10.22VNQWSPTNHH958 pKa = 6.65DD959 pKa = 3.27AEE961 pKa = 4.62EE962 pKa = 4.07IFRR965 pKa = 11.84VRR967 pKa = 11.84MVDD970 pKa = 3.26TTFSLIKK977 pKa = 10.4SNPYY981 pKa = 8.01YY982 pKa = 10.8HH983 pKa = 7.13EE984 pKa = 4.31VLTGISSPYY993 pKa = 10.21GLALRR998 pKa = 11.84PVPEE1002 pKa = 4.27EE1003 pKa = 3.89GRR1005 pKa = 11.84PSGSSS1010 pKa = 2.91
MM1 pKa = 7.48IRR3 pKa = 11.84LLINGPVPFGVTRR16 pKa = 11.84GAVNLSLINLQNALFSTSCVILRR39 pKa = 11.84KK40 pKa = 10.2YY41 pKa = 8.97EE42 pKa = 3.77VWTNLAKK49 pKa = 10.58RR50 pKa = 11.84HH51 pKa = 5.09GFAPAEE57 pKa = 4.23VIEE60 pKa = 4.32ASALGAYY67 pKa = 9.18VVVDD71 pKa = 3.86PMNKK75 pKa = 9.66SSLLALSPMAYY86 pKa = 9.59QKK88 pKa = 11.44LLGQTMSTEE97 pKa = 4.0NLSLILLLSPLDD109 pKa = 3.64EE110 pKa = 4.89LEE112 pKa = 4.23RR113 pKa = 11.84SAAEE117 pKa = 4.09ASPEE121 pKa = 4.05ATKK124 pKa = 10.8SPPLKK129 pKa = 10.69GFVNKK134 pKa = 10.18ISLDD138 pKa = 3.6LEE140 pKa = 4.09RR141 pKa = 11.84LKK143 pKa = 11.05EE144 pKa = 4.22RR145 pKa = 11.84IQTGRR150 pKa = 11.84LTKK153 pKa = 10.21GVRR156 pKa = 11.84WPSWSLGYY164 pKa = 9.67PFQGYY169 pKa = 9.29QGLGCRR175 pKa = 11.84LSDD178 pKa = 3.4GRR180 pKa = 11.84LILSRR185 pKa = 11.84PSEE188 pKa = 4.05FSAIFQRR195 pKa = 11.84WASRR199 pKa = 11.84LHH201 pKa = 5.58FWSGSSRR208 pKa = 11.84GRR210 pKa = 11.84RR211 pKa = 11.84FRR213 pKa = 11.84SSLWVVTNHH222 pKa = 5.85LQTVLKK228 pKa = 10.71HH229 pKa = 5.53SGPYY233 pKa = 10.23GLALYY238 pKa = 10.25LKK240 pKa = 9.8ASYY243 pKa = 10.66NIVLRR248 pKa = 11.84YY249 pKa = 9.6VAGQPFEE256 pKa = 4.58HH257 pKa = 7.22AFTQCGVPVQLQNGLPKK274 pKa = 10.4FLPRR278 pKa = 11.84EE279 pKa = 3.69WRR281 pKa = 11.84VAIRR285 pKa = 11.84SGSPRR290 pKa = 11.84LLRR293 pKa = 11.84IVLSLLYY300 pKa = 10.18SYY302 pKa = 11.16KK303 pKa = 10.53GFKK306 pKa = 8.19ATKK309 pKa = 8.28PKK311 pKa = 10.09PVAEE315 pKa = 3.9IFKK318 pKa = 10.25NISLAAHH325 pKa = 5.9QPDD328 pKa = 5.36DD329 pKa = 4.43IIAFSKK335 pKa = 10.84FCFEE339 pKa = 4.04FLKK342 pKa = 11.0NRR344 pKa = 11.84ISPISEE350 pKa = 4.05GEE352 pKa = 4.17LNPPSLPASLKK363 pKa = 10.54AGPNGPLSALAYY375 pKa = 10.34GADD378 pKa = 3.43AYY380 pKa = 10.9AWIQRR385 pKa = 11.84EE386 pKa = 4.13SAPYY390 pKa = 8.84QPSVPGPLEE399 pKa = 4.03WVQHH403 pKa = 5.8FGLDD407 pKa = 2.98KK408 pKa = 10.24WEE410 pKa = 4.29KK411 pKa = 10.44KK412 pKa = 9.31FRR414 pKa = 11.84AYY416 pKa = 10.95SDD418 pKa = 3.15WFLKK422 pKa = 10.62RR423 pKa = 11.84GFNHH427 pKa = 5.57QFNVTFPPPTRR438 pKa = 11.84KK439 pKa = 10.17GFGVLKK445 pKa = 10.54LKK447 pKa = 10.41IALGLNVPEE456 pKa = 4.78SKK458 pKa = 10.72SIALNPKK465 pKa = 8.92FAFHH469 pKa = 7.04RR470 pKa = 11.84RR471 pKa = 11.84TVSYY475 pKa = 10.0TPTRR479 pKa = 11.84VEE481 pKa = 4.24DD482 pKa = 4.27VEE484 pKa = 5.01ALQCLGLLLPVGRR497 pKa = 11.84VHH499 pKa = 6.76VLWEE503 pKa = 4.0AAGKK507 pKa = 7.82TRR509 pKa = 11.84VIAMMDD515 pKa = 3.94GLRR518 pKa = 11.84QALLRR523 pKa = 11.84PVHH526 pKa = 6.15LAMFRR531 pKa = 11.84RR532 pKa = 11.84IDD534 pKa = 3.83KK535 pKa = 10.84IFGFCSGLYY544 pKa = 9.18DD545 pKa = 3.08QSGAVRR551 pKa = 11.84SFAALNNRR559 pKa = 11.84EE560 pKa = 4.09VFSYY564 pKa = 10.47DD565 pKa = 2.51ISAATDD571 pKa = 3.86TIPYY575 pKa = 9.76QLYY578 pKa = 8.91FPLMEE583 pKa = 4.55FLLGKK588 pKa = 8.87TGAVLWNHH596 pKa = 5.95MVRR599 pKa = 11.84GHH601 pKa = 6.33PFIHH605 pKa = 6.93LEE607 pKa = 4.33GGDD610 pKa = 3.92EE611 pKa = 3.95PWAQRR616 pKa = 11.84LAGSSSYY623 pKa = 10.82YY624 pKa = 9.26YY625 pKa = 10.61EE626 pKa = 4.96RR627 pKa = 11.84GQPMGGYY634 pKa = 9.96SSFAALDD641 pKa = 3.84MLHH644 pKa = 6.74HH645 pKa = 7.09LIVQYY650 pKa = 10.96SAFEE654 pKa = 4.12AGYY657 pKa = 8.37DD658 pKa = 3.78TLNKK662 pKa = 9.88TFDD665 pKa = 3.26NYY667 pKa = 10.83RR668 pKa = 11.84ILGDD672 pKa = 3.74DD673 pKa = 3.63VVIGDD678 pKa = 3.48VRR680 pKa = 11.84VAKK683 pKa = 9.91TYY685 pKa = 11.11LNFMRR690 pKa = 11.84TWNIPISEE698 pKa = 4.25NKK700 pKa = 10.33SLVSSKK706 pKa = 11.01GVFQFLSEE714 pKa = 4.01VFRR717 pKa = 11.84GGEE720 pKa = 4.15CLSPLSFRR728 pKa = 11.84ADD730 pKa = 3.64YY731 pKa = 10.43QSKK734 pKa = 8.02TFLEE738 pKa = 4.41RR739 pKa = 11.84FSFAATAVSRR749 pKa = 11.84GWSSGLAMEE758 pKa = 5.02GFGRR762 pKa = 11.84LILSKK767 pKa = 10.89DD768 pKa = 3.27QFLSEE773 pKa = 3.22IHH775 pKa = 7.07RR776 pKa = 11.84YY777 pKa = 8.66RR778 pKa = 11.84NRR780 pKa = 11.84RR781 pKa = 11.84PSAFIYY787 pKa = 10.59SLLFALVTFAGEE799 pKa = 4.18AGWALGSWLHH809 pKa = 5.92NVVAHH814 pKa = 7.25DD815 pKa = 4.96PFPQLLSNKK824 pKa = 7.69EE825 pKa = 4.09LEE827 pKa = 4.48SVEE830 pKa = 4.22SKK832 pKa = 11.2NFVFTILRR840 pKa = 11.84QILFTEE846 pKa = 4.72AGRR849 pKa = 11.84IHH851 pKa = 6.98EE852 pKa = 4.19LQKK855 pKa = 11.11AEE857 pKa = 4.16SGVNQTRR864 pKa = 11.84QFLDD868 pKa = 3.71FGSIEE873 pKa = 3.91FHH875 pKa = 6.95NDD877 pKa = 2.19IEE879 pKa = 4.72NDD881 pKa = 3.43VDD883 pKa = 4.9SLWLLARR890 pKa = 11.84EE891 pKa = 4.43GKK893 pKa = 8.11TLLEE897 pKa = 4.55SADD900 pKa = 3.74VTPDD904 pKa = 2.7WVTSAFSYY912 pKa = 9.98LRR914 pKa = 11.84KK915 pKa = 9.97VSGIIPRR922 pKa = 11.84LSATGVVHH930 pKa = 6.72PLDD933 pKa = 3.88YY934 pKa = 10.98LVLMKK939 pKa = 8.87TTEE942 pKa = 3.97DD943 pKa = 3.29RR944 pKa = 11.84NNIHH948 pKa = 7.14KK949 pKa = 10.22VNQWSPTNHH958 pKa = 6.65DD959 pKa = 3.27AEE961 pKa = 4.62EE962 pKa = 4.07IFRR965 pKa = 11.84VRR967 pKa = 11.84MVDD970 pKa = 3.26TTFSLIKK977 pKa = 10.4SNPYY981 pKa = 8.01YY982 pKa = 10.8HH983 pKa = 7.13EE984 pKa = 4.31VLTGISSPYY993 pKa = 10.21GLALRR998 pKa = 11.84PVPEE1002 pKa = 4.27EE1003 pKa = 3.89GRR1005 pKa = 11.84PSGSSS1010 pKa = 2.91
Molecular weight: 113.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1010 |
1010 |
1010 |
1010.0 |
113.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.525 ± 0.0 | 0.693 ± 0.0 |
3.465 ± 0.0 | 5.248 ± 0.0 |
6.04 ± 0.0 | 7.129 ± 0.0 |
2.475 ± 0.0 | 4.653 ± 0.0 |
4.554 ± 0.0 | 12.079 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.485 ± 0.0 | 3.861 ± 0.0 |
5.743 ± 0.0 | 3.267 ± 0.0 |
6.535 ± 0.0 | 9.109 ± 0.0 |
4.059 ± 0.0 | 6.436 ± 0.0 |
2.079 ± 0.0 | 3.564 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
