
Porcine rotavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus C
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
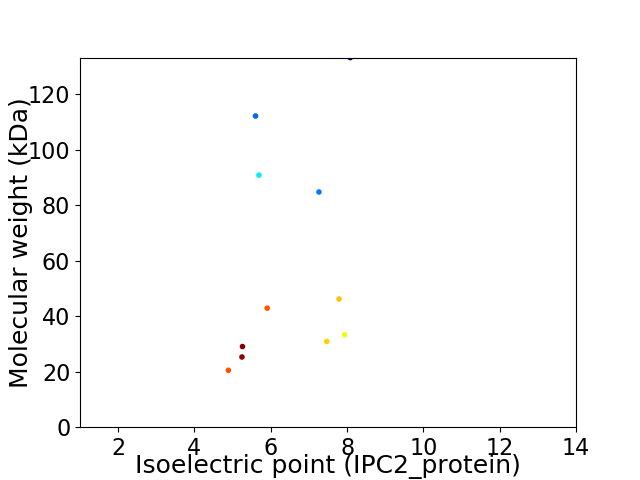
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
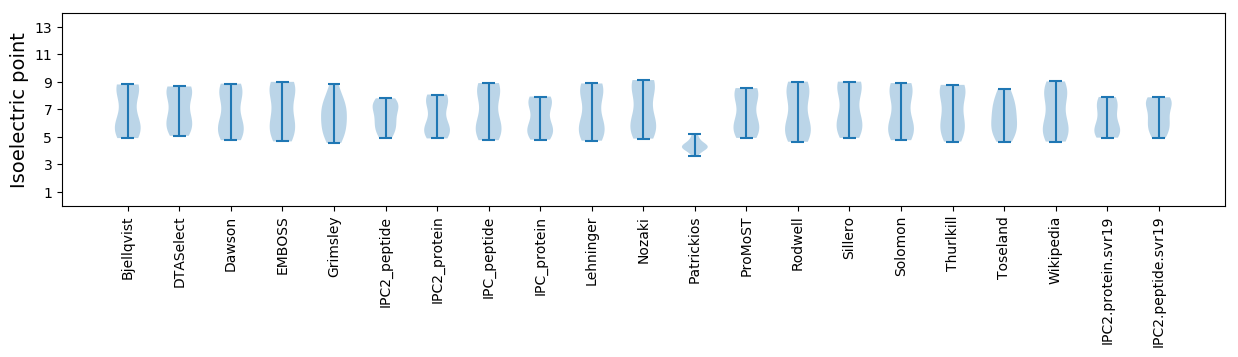
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K2SR53|A0A0K2SR53_9REOV NSP2 OS=Porcine rotavirus OX=10913 PE=4 SV=1
MM1 pKa = 7.57SDD3 pKa = 3.24VPRR6 pKa = 11.84FDD8 pKa = 4.66LRR10 pKa = 11.84SKK12 pKa = 10.93RR13 pKa = 11.84KK14 pKa = 5.54TQKK17 pKa = 9.38KK18 pKa = 8.25QKK20 pKa = 10.39VDD22 pKa = 3.28IFDD25 pKa = 4.36EE26 pKa = 4.48DD27 pKa = 4.13DD28 pKa = 3.99KK29 pKa = 11.62EE30 pKa = 5.11SSIQVDD36 pKa = 4.07GEE38 pKa = 4.3SDD40 pKa = 3.61SLASADD46 pKa = 3.44TSSNHH51 pKa = 6.35SYY53 pKa = 11.12EE54 pKa = 5.07DD55 pKa = 3.49YY56 pKa = 11.26SKK58 pKa = 11.08AYY60 pKa = 9.65KK61 pKa = 10.4DD62 pKa = 3.68LTLEE66 pKa = 4.42TPVEE70 pKa = 3.98QPIGDD75 pKa = 4.41DD76 pKa = 3.71DD77 pKa = 4.14QVDD80 pKa = 4.01SVSTILEE87 pKa = 4.3SKK89 pKa = 10.42VDD91 pKa = 3.59NSWYY95 pKa = 10.22DD96 pKa = 3.04KK97 pKa = 10.07TVEE100 pKa = 4.0VEE102 pKa = 4.13EE103 pKa = 4.78RR104 pKa = 11.84VQKK107 pKa = 9.76PKK109 pKa = 11.08RR110 pKa = 11.84MTKK113 pKa = 10.02QKK115 pKa = 10.54AQQIDD120 pKa = 4.28EE121 pKa = 4.42DD122 pKa = 4.24TSDD125 pKa = 5.23LKK127 pKa = 11.36LQLAQLSLRR136 pKa = 11.84IQKK139 pKa = 8.92MEE141 pKa = 3.84SEE143 pKa = 4.5TRR145 pKa = 11.84AKK147 pKa = 10.04TIDD150 pKa = 3.53SAYY153 pKa = 9.19HH154 pKa = 5.64TIITQAEE161 pKa = 4.42NLTTPQKK168 pKa = 10.61KK169 pKa = 10.11SLIAAIIATMRR180 pKa = 3.55
MM1 pKa = 7.57SDD3 pKa = 3.24VPRR6 pKa = 11.84FDD8 pKa = 4.66LRR10 pKa = 11.84SKK12 pKa = 10.93RR13 pKa = 11.84KK14 pKa = 5.54TQKK17 pKa = 9.38KK18 pKa = 8.25QKK20 pKa = 10.39VDD22 pKa = 3.28IFDD25 pKa = 4.36EE26 pKa = 4.48DD27 pKa = 4.13DD28 pKa = 3.99KK29 pKa = 11.62EE30 pKa = 5.11SSIQVDD36 pKa = 4.07GEE38 pKa = 4.3SDD40 pKa = 3.61SLASADD46 pKa = 3.44TSSNHH51 pKa = 6.35SYY53 pKa = 11.12EE54 pKa = 5.07DD55 pKa = 3.49YY56 pKa = 11.26SKK58 pKa = 11.08AYY60 pKa = 9.65KK61 pKa = 10.4DD62 pKa = 3.68LTLEE66 pKa = 4.42TPVEE70 pKa = 3.98QPIGDD75 pKa = 4.41DD76 pKa = 3.71DD77 pKa = 4.14QVDD80 pKa = 4.01SVSTILEE87 pKa = 4.3SKK89 pKa = 10.42VDD91 pKa = 3.59NSWYY95 pKa = 10.22DD96 pKa = 3.04KK97 pKa = 10.07TVEE100 pKa = 4.0VEE102 pKa = 4.13EE103 pKa = 4.78RR104 pKa = 11.84VQKK107 pKa = 9.76PKK109 pKa = 11.08RR110 pKa = 11.84MTKK113 pKa = 10.02QKK115 pKa = 10.54AQQIDD120 pKa = 4.28EE121 pKa = 4.42DD122 pKa = 4.24TSDD125 pKa = 5.23LKK127 pKa = 11.36LQLAQLSLRR136 pKa = 11.84IQKK139 pKa = 8.92MEE141 pKa = 3.84SEE143 pKa = 4.5TRR145 pKa = 11.84AKK147 pKa = 10.04TIDD150 pKa = 3.53SAYY153 pKa = 9.19HH154 pKa = 5.64TIITQAEE161 pKa = 4.42NLTTPQKK168 pKa = 10.61KK169 pKa = 10.11SLIAAIIATMRR180 pKa = 3.55
Molecular weight: 20.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K2SRD6|A0A0K2SRD6_9REOV NSP3 OS=Porcine rotavirus OX=10913 PE=4 SV=1
MM1 pKa = 8.15DD2 pKa = 5.07GANYY6 pKa = 9.77LAWLARR12 pKa = 11.84DD13 pKa = 3.39IVRR16 pKa = 11.84NLSYY20 pKa = 10.23TALVYY25 pKa = 10.85NNPKK29 pKa = 9.62VAIVTLQDD37 pKa = 3.37SKK39 pKa = 11.87EE40 pKa = 3.91EE41 pKa = 4.22FISFEE46 pKa = 4.33KK47 pKa = 9.68EE48 pKa = 3.49QKK50 pKa = 9.31TPSEE54 pKa = 4.74LIAEE58 pKa = 4.24IQNIVEE64 pKa = 4.5SNLSIEE70 pKa = 4.82DD71 pKa = 4.07KK72 pKa = 10.92INNLLTIRR80 pKa = 11.84YY81 pKa = 8.36ISVYY85 pKa = 10.67VDD87 pKa = 3.43DD88 pKa = 5.87KK89 pKa = 10.99SDD91 pKa = 3.17KK92 pKa = 10.94RR93 pKa = 11.84EE94 pKa = 4.08IVSKK98 pKa = 10.9LLDD101 pKa = 4.29KK102 pKa = 10.69IIKK105 pKa = 8.25TLEE108 pKa = 4.02NKK110 pKa = 9.9VSISEE115 pKa = 4.23TLKK118 pKa = 10.92KK119 pKa = 10.81CIDD122 pKa = 4.62DD123 pKa = 3.62IRR125 pKa = 11.84NEE127 pKa = 4.01AKK129 pKa = 9.61SWRR132 pKa = 11.84IKK134 pKa = 10.64NSSSFKK140 pKa = 10.09NYY142 pKa = 9.99HH143 pKa = 5.38YY144 pKa = 10.73NQLVSDD150 pKa = 4.36FIKK153 pKa = 10.99YY154 pKa = 10.41NEE156 pKa = 3.92FEE158 pKa = 4.06ILEE161 pKa = 4.75DD162 pKa = 3.52EE163 pKa = 5.68DD164 pKa = 3.83EE165 pKa = 4.54YY166 pKa = 11.38RR167 pKa = 11.84WKK169 pKa = 10.91SDD171 pKa = 3.13TLQGLSPNYY180 pKa = 9.03NHH182 pKa = 6.95RR183 pKa = 11.84THH185 pKa = 6.5TLISSIIFAVQSRR198 pKa = 11.84MLRR201 pKa = 11.84YY202 pKa = 9.56NDD204 pKa = 3.43EE205 pKa = 4.06QLEE208 pKa = 4.3VLLFMFNKK216 pKa = 9.44IRR218 pKa = 11.84EE219 pKa = 4.49CYY221 pKa = 9.17NDD223 pKa = 4.35GYY225 pKa = 11.58LEE227 pKa = 4.31LLPNRR232 pKa = 11.84KK233 pKa = 8.54WSHH236 pKa = 5.4TIKK239 pKa = 10.85DD240 pKa = 3.3LRR242 pKa = 11.84TNKK245 pKa = 9.68KK246 pKa = 9.58IKK248 pKa = 9.6MYY250 pKa = 9.16SAKK253 pKa = 9.95IIHH256 pKa = 6.37ATCAMISILHH266 pKa = 7.08ADD268 pKa = 4.29PIDD271 pKa = 4.2PYY273 pKa = 11.25FLAQIVTTFEE283 pKa = 4.59EE284 pKa = 4.94IPAHH288 pKa = 6.13AAKK291 pKa = 10.27QLSSPMTLYY300 pKa = 10.45IGVAQIKK307 pKa = 9.84SNIVISTQRR316 pKa = 11.84AAEE319 pKa = 4.15SVASIKK325 pKa = 10.77PNVSRR330 pKa = 11.84LDD332 pKa = 3.64EE333 pKa = 4.35SQVAEE338 pKa = 4.29WNEE341 pKa = 3.78EE342 pKa = 3.39MDD344 pKa = 3.77EE345 pKa = 4.19YY346 pKa = 11.08HH347 pKa = 6.73FKK349 pKa = 11.17SSILVNMMKK358 pKa = 10.23TNIYY362 pKa = 9.64DD363 pKa = 3.85VPVHH367 pKa = 5.62TFYY370 pKa = 11.2KK371 pKa = 10.33IFNCFSATFHH381 pKa = 6.21TGHH384 pKa = 6.89RR385 pKa = 11.84VDD387 pKa = 4.38NPQDD391 pKa = 3.96AIEE394 pKa = 4.35AQVKK398 pKa = 9.4VEE400 pKa = 3.92YY401 pKa = 10.13TSDD404 pKa = 3.3INRR407 pKa = 11.84EE408 pKa = 4.04MYY410 pKa = 9.66DD411 pKa = 2.82QYY413 pKa = 11.92YY414 pKa = 10.45FLLKK418 pKa = 10.66RR419 pKa = 11.84MLTDD423 pKa = 3.07QFAEE427 pKa = 4.23YY428 pKa = 10.49AEE430 pKa = 4.3EE431 pKa = 4.15MFHH434 pKa = 7.21KK435 pKa = 10.7YY436 pKa = 10.57NSDD439 pKa = 3.12VTAEE443 pKa = 4.32SLASMSNSSNGYY455 pKa = 7.81SRR457 pKa = 11.84SVKK460 pKa = 10.62FIDD463 pKa = 5.01RR464 pKa = 11.84EE465 pKa = 4.01IKK467 pKa = 6.98TTKK470 pKa = 10.6KK471 pKa = 9.58MLHH474 pKa = 6.67LDD476 pKa = 3.88DD477 pKa = 6.17DD478 pKa = 4.61LSKK481 pKa = 11.35DD482 pKa = 4.18LDD484 pKa = 3.72FTNIGEE490 pKa = 4.39QISKK494 pKa = 9.63GIPMGTRR501 pKa = 11.84NVPARR506 pKa = 11.84QTRR509 pKa = 11.84GIFILNWQVAAIQHH523 pKa = 6.29TIAEE527 pKa = 4.27FLYY530 pKa = 10.6KK531 pKa = 9.96KK532 pKa = 10.11AKK534 pKa = 9.99KK535 pKa = 10.15GGFGATFAEE544 pKa = 4.84AYY546 pKa = 8.25VAKK549 pKa = 10.32AATLTYY555 pKa = 10.43GILAEE560 pKa = 4.34ATSKK564 pKa = 11.11ADD566 pKa = 3.65QLILYY571 pKa = 8.59TDD573 pKa = 3.83VSQWDD578 pKa = 3.7ASQHH582 pKa = 4.09NTEE585 pKa = 4.88PYY587 pKa = 9.9RR588 pKa = 11.84SAIINAIKK596 pKa = 10.07EE597 pKa = 3.89ARR599 pKa = 11.84TKK601 pKa = 11.05YY602 pKa = 10.15KK603 pKa = 10.01ISYY606 pKa = 7.25EE607 pKa = 3.97QEE609 pKa = 3.97PKK611 pKa = 10.14VLGMNVLDD619 pKa = 5.15KK620 pKa = 10.67MIEE623 pKa = 3.92IQEE626 pKa = 4.22ALLNSNLLVEE636 pKa = 4.61SQGSKK641 pKa = 10.4RR642 pKa = 11.84PPLKK646 pKa = 9.94IRR648 pKa = 11.84YY649 pKa = 8.39HH650 pKa = 5.84GVASGEE656 pKa = 4.15KK657 pKa = 6.55TTKK660 pKa = 10.0IGNSFANVALITTVFNSLTNSLPSIRR686 pKa = 11.84VNHH689 pKa = 5.66MRR691 pKa = 11.84VDD693 pKa = 3.56GDD695 pKa = 4.26DD696 pKa = 3.95NVVTMYY702 pKa = 9.86TADD705 pKa = 4.92PINVVQEE712 pKa = 4.32AIKK715 pKa = 9.92TKK717 pKa = 10.38YY718 pKa = 10.19KK719 pKa = 11.05DD720 pKa = 2.91MNARR724 pKa = 11.84VKK726 pKa = 10.79ALASYY731 pKa = 9.46TGLEE735 pKa = 3.81MAKK738 pKa = 10.13RR739 pKa = 11.84FIIGGKK745 pKa = 8.6IFEE748 pKa = 4.99RR749 pKa = 11.84GAISIFTAEE758 pKa = 4.3RR759 pKa = 11.84PYY761 pKa = 10.8GTDD764 pKa = 2.76QSVQSTTGSLIYY776 pKa = 10.34SAAVNAYY783 pKa = 9.61RR784 pKa = 11.84GFGDD788 pKa = 4.44GYY790 pKa = 10.86LEE792 pKa = 4.44FMLDD796 pKa = 3.37VLTPPSASIRR806 pKa = 11.84VTGRR810 pKa = 11.84LRR812 pKa = 11.84SLLSPVTLFSTGPISFEE829 pKa = 3.43ITPYY833 pKa = 10.91GLGGRR838 pKa = 11.84MRR840 pKa = 11.84FFQINRR846 pKa = 11.84RR847 pKa = 11.84NMEE850 pKa = 4.02LFKK853 pKa = 10.8ILTSSLSISIEE864 pKa = 3.87PEE866 pKa = 3.66EE867 pKa = 4.09VKK869 pKa = 10.46QYY871 pKa = 11.88SMTPQFKK878 pKa = 10.53ARR880 pKa = 11.84TDD882 pKa = 3.32RR883 pKa = 11.84MIEE886 pKa = 4.06SVRR889 pKa = 11.84IAMKK893 pKa = 10.56SEE895 pKa = 3.74AKK897 pKa = 10.2IITQILVDD905 pKa = 4.0KK906 pKa = 10.27EE907 pKa = 4.11RR908 pKa = 11.84QKK910 pKa = 9.96TLGVPNVATTKK921 pKa = 10.27NRR923 pKa = 11.84QQVEE927 pKa = 4.42KK928 pKa = 10.84ARR930 pKa = 11.84KK931 pKa = 6.81TLSAPKK937 pKa = 9.81EE938 pKa = 4.04VLKK941 pKa = 10.83KK942 pKa = 9.49VSKK945 pKa = 10.46YY946 pKa = 8.33YY947 pKa = 9.95PEE949 pKa = 5.29EE950 pKa = 3.73IFHH953 pKa = 6.33YY954 pKa = 10.24IISNSKK960 pKa = 8.97LHH962 pKa = 6.96KK963 pKa = 9.89SQNDD967 pKa = 3.45EE968 pKa = 3.97KK969 pKa = 11.24VQVYY973 pKa = 8.38MNNSTMIKK981 pKa = 9.62QLQRR985 pKa = 11.84QLGVRR990 pKa = 11.84VAEE993 pKa = 4.9GSHH996 pKa = 5.16LTKK999 pKa = 9.5PTNTLFKK1006 pKa = 10.45LVEE1009 pKa = 3.9KK1010 pKa = 10.28HH1011 pKa = 6.31SPIKK1015 pKa = 10.33ISPSDD1020 pKa = 3.71LVKK1023 pKa = 10.91HH1024 pKa = 5.3SEE1026 pKa = 4.17KK1027 pKa = 10.73YY1028 pKa = 10.41DD1029 pKa = 3.51LSTLQGKK1036 pKa = 9.46KK1037 pKa = 10.15KK1038 pKa = 10.14FLYY1041 pKa = 10.59DD1042 pKa = 3.72LGISGSEE1049 pKa = 3.42LRR1051 pKa = 11.84FYY1053 pKa = 11.3LNSKK1057 pKa = 10.47LLMHH1061 pKa = 7.24DD1062 pKa = 4.58LLLAKK1067 pKa = 10.02YY1068 pKa = 9.39DD1069 pKa = 3.98KK1070 pKa = 10.88LYY1072 pKa = 9.67EE1073 pKa = 4.3APGFGATQLNSLPLDD1088 pKa = 3.64LTAAEE1093 pKa = 4.75KK1094 pKa = 10.88VFSVKK1099 pKa = 10.31VNMPSSYY1106 pKa = 10.65YY1107 pKa = 10.48EE1108 pKa = 4.14LMMLILLYY1116 pKa = 10.42EE1117 pKa = 4.09YY1118 pKa = 11.42VNFVMFTGQTFTCICLPEE1136 pKa = 4.12TTIQSANIAKK1146 pKa = 10.12NVMKK1150 pKa = 10.15MIDD1153 pKa = 4.2NIQLDD1158 pKa = 4.16SVSFQDD1164 pKa = 4.41VIYY1167 pKa = 10.98
MM1 pKa = 8.15DD2 pKa = 5.07GANYY6 pKa = 9.77LAWLARR12 pKa = 11.84DD13 pKa = 3.39IVRR16 pKa = 11.84NLSYY20 pKa = 10.23TALVYY25 pKa = 10.85NNPKK29 pKa = 9.62VAIVTLQDD37 pKa = 3.37SKK39 pKa = 11.87EE40 pKa = 3.91EE41 pKa = 4.22FISFEE46 pKa = 4.33KK47 pKa = 9.68EE48 pKa = 3.49QKK50 pKa = 9.31TPSEE54 pKa = 4.74LIAEE58 pKa = 4.24IQNIVEE64 pKa = 4.5SNLSIEE70 pKa = 4.82DD71 pKa = 4.07KK72 pKa = 10.92INNLLTIRR80 pKa = 11.84YY81 pKa = 8.36ISVYY85 pKa = 10.67VDD87 pKa = 3.43DD88 pKa = 5.87KK89 pKa = 10.99SDD91 pKa = 3.17KK92 pKa = 10.94RR93 pKa = 11.84EE94 pKa = 4.08IVSKK98 pKa = 10.9LLDD101 pKa = 4.29KK102 pKa = 10.69IIKK105 pKa = 8.25TLEE108 pKa = 4.02NKK110 pKa = 9.9VSISEE115 pKa = 4.23TLKK118 pKa = 10.92KK119 pKa = 10.81CIDD122 pKa = 4.62DD123 pKa = 3.62IRR125 pKa = 11.84NEE127 pKa = 4.01AKK129 pKa = 9.61SWRR132 pKa = 11.84IKK134 pKa = 10.64NSSSFKK140 pKa = 10.09NYY142 pKa = 9.99HH143 pKa = 5.38YY144 pKa = 10.73NQLVSDD150 pKa = 4.36FIKK153 pKa = 10.99YY154 pKa = 10.41NEE156 pKa = 3.92FEE158 pKa = 4.06ILEE161 pKa = 4.75DD162 pKa = 3.52EE163 pKa = 5.68DD164 pKa = 3.83EE165 pKa = 4.54YY166 pKa = 11.38RR167 pKa = 11.84WKK169 pKa = 10.91SDD171 pKa = 3.13TLQGLSPNYY180 pKa = 9.03NHH182 pKa = 6.95RR183 pKa = 11.84THH185 pKa = 6.5TLISSIIFAVQSRR198 pKa = 11.84MLRR201 pKa = 11.84YY202 pKa = 9.56NDD204 pKa = 3.43EE205 pKa = 4.06QLEE208 pKa = 4.3VLLFMFNKK216 pKa = 9.44IRR218 pKa = 11.84EE219 pKa = 4.49CYY221 pKa = 9.17NDD223 pKa = 4.35GYY225 pKa = 11.58LEE227 pKa = 4.31LLPNRR232 pKa = 11.84KK233 pKa = 8.54WSHH236 pKa = 5.4TIKK239 pKa = 10.85DD240 pKa = 3.3LRR242 pKa = 11.84TNKK245 pKa = 9.68KK246 pKa = 9.58IKK248 pKa = 9.6MYY250 pKa = 9.16SAKK253 pKa = 9.95IIHH256 pKa = 6.37ATCAMISILHH266 pKa = 7.08ADD268 pKa = 4.29PIDD271 pKa = 4.2PYY273 pKa = 11.25FLAQIVTTFEE283 pKa = 4.59EE284 pKa = 4.94IPAHH288 pKa = 6.13AAKK291 pKa = 10.27QLSSPMTLYY300 pKa = 10.45IGVAQIKK307 pKa = 9.84SNIVISTQRR316 pKa = 11.84AAEE319 pKa = 4.15SVASIKK325 pKa = 10.77PNVSRR330 pKa = 11.84LDD332 pKa = 3.64EE333 pKa = 4.35SQVAEE338 pKa = 4.29WNEE341 pKa = 3.78EE342 pKa = 3.39MDD344 pKa = 3.77EE345 pKa = 4.19YY346 pKa = 11.08HH347 pKa = 6.73FKK349 pKa = 11.17SSILVNMMKK358 pKa = 10.23TNIYY362 pKa = 9.64DD363 pKa = 3.85VPVHH367 pKa = 5.62TFYY370 pKa = 11.2KK371 pKa = 10.33IFNCFSATFHH381 pKa = 6.21TGHH384 pKa = 6.89RR385 pKa = 11.84VDD387 pKa = 4.38NPQDD391 pKa = 3.96AIEE394 pKa = 4.35AQVKK398 pKa = 9.4VEE400 pKa = 3.92YY401 pKa = 10.13TSDD404 pKa = 3.3INRR407 pKa = 11.84EE408 pKa = 4.04MYY410 pKa = 9.66DD411 pKa = 2.82QYY413 pKa = 11.92YY414 pKa = 10.45FLLKK418 pKa = 10.66RR419 pKa = 11.84MLTDD423 pKa = 3.07QFAEE427 pKa = 4.23YY428 pKa = 10.49AEE430 pKa = 4.3EE431 pKa = 4.15MFHH434 pKa = 7.21KK435 pKa = 10.7YY436 pKa = 10.57NSDD439 pKa = 3.12VTAEE443 pKa = 4.32SLASMSNSSNGYY455 pKa = 7.81SRR457 pKa = 11.84SVKK460 pKa = 10.62FIDD463 pKa = 5.01RR464 pKa = 11.84EE465 pKa = 4.01IKK467 pKa = 6.98TTKK470 pKa = 10.6KK471 pKa = 9.58MLHH474 pKa = 6.67LDD476 pKa = 3.88DD477 pKa = 6.17DD478 pKa = 4.61LSKK481 pKa = 11.35DD482 pKa = 4.18LDD484 pKa = 3.72FTNIGEE490 pKa = 4.39QISKK494 pKa = 9.63GIPMGTRR501 pKa = 11.84NVPARR506 pKa = 11.84QTRR509 pKa = 11.84GIFILNWQVAAIQHH523 pKa = 6.29TIAEE527 pKa = 4.27FLYY530 pKa = 10.6KK531 pKa = 9.96KK532 pKa = 10.11AKK534 pKa = 9.99KK535 pKa = 10.15GGFGATFAEE544 pKa = 4.84AYY546 pKa = 8.25VAKK549 pKa = 10.32AATLTYY555 pKa = 10.43GILAEE560 pKa = 4.34ATSKK564 pKa = 11.11ADD566 pKa = 3.65QLILYY571 pKa = 8.59TDD573 pKa = 3.83VSQWDD578 pKa = 3.7ASQHH582 pKa = 4.09NTEE585 pKa = 4.88PYY587 pKa = 9.9RR588 pKa = 11.84SAIINAIKK596 pKa = 10.07EE597 pKa = 3.89ARR599 pKa = 11.84TKK601 pKa = 11.05YY602 pKa = 10.15KK603 pKa = 10.01ISYY606 pKa = 7.25EE607 pKa = 3.97QEE609 pKa = 3.97PKK611 pKa = 10.14VLGMNVLDD619 pKa = 5.15KK620 pKa = 10.67MIEE623 pKa = 3.92IQEE626 pKa = 4.22ALLNSNLLVEE636 pKa = 4.61SQGSKK641 pKa = 10.4RR642 pKa = 11.84PPLKK646 pKa = 9.94IRR648 pKa = 11.84YY649 pKa = 8.39HH650 pKa = 5.84GVASGEE656 pKa = 4.15KK657 pKa = 6.55TTKK660 pKa = 10.0IGNSFANVALITTVFNSLTNSLPSIRR686 pKa = 11.84VNHH689 pKa = 5.66MRR691 pKa = 11.84VDD693 pKa = 3.56GDD695 pKa = 4.26DD696 pKa = 3.95NVVTMYY702 pKa = 9.86TADD705 pKa = 4.92PINVVQEE712 pKa = 4.32AIKK715 pKa = 9.92TKK717 pKa = 10.38YY718 pKa = 10.19KK719 pKa = 11.05DD720 pKa = 2.91MNARR724 pKa = 11.84VKK726 pKa = 10.79ALASYY731 pKa = 9.46TGLEE735 pKa = 3.81MAKK738 pKa = 10.13RR739 pKa = 11.84FIIGGKK745 pKa = 8.6IFEE748 pKa = 4.99RR749 pKa = 11.84GAISIFTAEE758 pKa = 4.3RR759 pKa = 11.84PYY761 pKa = 10.8GTDD764 pKa = 2.76QSVQSTTGSLIYY776 pKa = 10.34SAAVNAYY783 pKa = 9.61RR784 pKa = 11.84GFGDD788 pKa = 4.44GYY790 pKa = 10.86LEE792 pKa = 4.44FMLDD796 pKa = 3.37VLTPPSASIRR806 pKa = 11.84VTGRR810 pKa = 11.84LRR812 pKa = 11.84SLLSPVTLFSTGPISFEE829 pKa = 3.43ITPYY833 pKa = 10.91GLGGRR838 pKa = 11.84MRR840 pKa = 11.84FFQINRR846 pKa = 11.84RR847 pKa = 11.84NMEE850 pKa = 4.02LFKK853 pKa = 10.8ILTSSLSISIEE864 pKa = 3.87PEE866 pKa = 3.66EE867 pKa = 4.09VKK869 pKa = 10.46QYY871 pKa = 11.88SMTPQFKK878 pKa = 10.53ARR880 pKa = 11.84TDD882 pKa = 3.32RR883 pKa = 11.84MIEE886 pKa = 4.06SVRR889 pKa = 11.84IAMKK893 pKa = 10.56SEE895 pKa = 3.74AKK897 pKa = 10.2IITQILVDD905 pKa = 4.0KK906 pKa = 10.27EE907 pKa = 4.11RR908 pKa = 11.84QKK910 pKa = 9.96TLGVPNVATTKK921 pKa = 10.27NRR923 pKa = 11.84QQVEE927 pKa = 4.42KK928 pKa = 10.84ARR930 pKa = 11.84KK931 pKa = 6.81TLSAPKK937 pKa = 9.81EE938 pKa = 4.04VLKK941 pKa = 10.83KK942 pKa = 9.49VSKK945 pKa = 10.46YY946 pKa = 8.33YY947 pKa = 9.95PEE949 pKa = 5.29EE950 pKa = 3.73IFHH953 pKa = 6.33YY954 pKa = 10.24IISNSKK960 pKa = 8.97LHH962 pKa = 6.96KK963 pKa = 9.89SQNDD967 pKa = 3.45EE968 pKa = 3.97KK969 pKa = 11.24VQVYY973 pKa = 8.38MNNSTMIKK981 pKa = 9.62QLQRR985 pKa = 11.84QLGVRR990 pKa = 11.84VAEE993 pKa = 4.9GSHH996 pKa = 5.16LTKK999 pKa = 9.5PTNTLFKK1006 pKa = 10.45LVEE1009 pKa = 3.9KK1010 pKa = 10.28HH1011 pKa = 6.31SPIKK1015 pKa = 10.33ISPSDD1020 pKa = 3.71LVKK1023 pKa = 10.91HH1024 pKa = 5.3SEE1026 pKa = 4.17KK1027 pKa = 10.73YY1028 pKa = 10.41DD1029 pKa = 3.51LSTLQGKK1036 pKa = 9.46KK1037 pKa = 10.15KK1038 pKa = 10.14FLYY1041 pKa = 10.59DD1042 pKa = 3.72LGISGSEE1049 pKa = 3.42LRR1051 pKa = 11.84FYY1053 pKa = 11.3LNSKK1057 pKa = 10.47LLMHH1061 pKa = 7.24DD1062 pKa = 4.58LLLAKK1067 pKa = 10.02YY1068 pKa = 9.39DD1069 pKa = 3.98KK1070 pKa = 10.88LYY1072 pKa = 9.67EE1073 pKa = 4.3APGFGATQLNSLPLDD1088 pKa = 3.64LTAAEE1093 pKa = 4.75KK1094 pKa = 10.88VFSVKK1099 pKa = 10.31VNMPSSYY1106 pKa = 10.65YY1107 pKa = 10.48EE1108 pKa = 4.14LMMLILLYY1116 pKa = 10.42EE1117 pKa = 4.09YY1118 pKa = 11.42VNFVMFTGQTFTCICLPEE1136 pKa = 4.12TTIQSANIAKK1146 pKa = 10.12NVMKK1150 pKa = 10.15MIDD1153 pKa = 4.2NIQLDD1158 pKa = 4.16SVSFQDD1164 pKa = 4.41VIYY1167 pKa = 10.98
Molecular weight: 133.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5693 |
180 |
1167 |
517.5 |
59.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.499 ± 0.48 | 1.265 ± 0.371 |
6.043 ± 0.477 | 6.253 ± 0.427 |
4.426 ± 0.315 | 3.882 ± 0.315 |
1.862 ± 0.26 | 7.992 ± 0.32 |
6.692 ± 0.61 | 8.221 ± 0.419 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.881 ± 0.251 | 6.569 ± 0.387 |
3.952 ± 0.379 | 3.9 ± 0.307 |
5.445 ± 0.358 | 7.096 ± 0.579 |
6.447 ± 0.477 | 6.148 ± 0.319 |
0.755 ± 0.119 | 3.671 ± 0.428 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
