
Ceratocystis polonica partitivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus; Ceratocystis resinifera virus 1
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
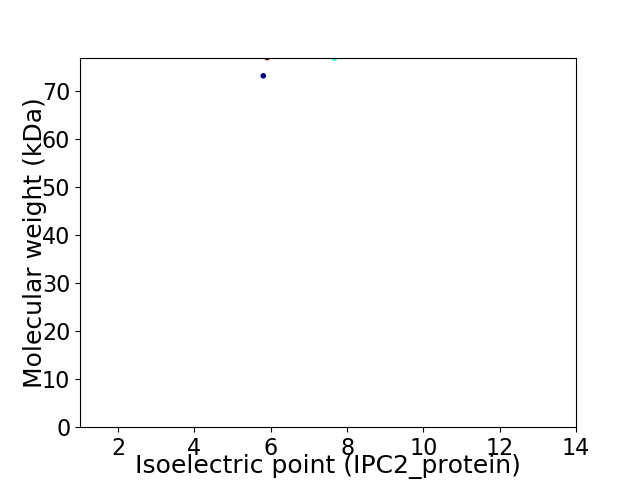
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6X9T6|Q6X9T6_9VIRU Putative RNA-dependent RNA polymerase OS=Ceratocystis polonica partitivirus OX=235434 GN=RDRP PE=4 SV=1
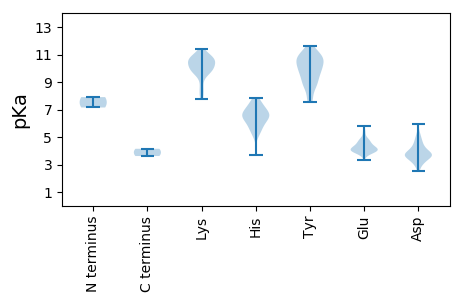
MM1 pKa = 7.15SHH3 pKa = 7.84DD4 pKa = 3.89ARR6 pKa = 11.84DD7 pKa = 3.75LQSEE11 pKa = 4.18ARR13 pKa = 11.84QEE15 pKa = 3.93VRR17 pKa = 11.84PSGLQPDD24 pKa = 3.99AHH26 pKa = 6.53SHH28 pKa = 5.66EE29 pKa = 5.37PIGLNNSIARR39 pKa = 11.84TAPSSSAGPGLTAPHH54 pKa = 6.94AGTRR58 pKa = 11.84SLGLPCKK65 pKa = 10.65GEE67 pKa = 4.03ASHH70 pKa = 7.34SSEE73 pKa = 4.0ASSLARR79 pKa = 11.84DD80 pKa = 3.02IDD82 pKa = 3.69LGRR85 pKa = 11.84NCSRR89 pKa = 11.84SSGEE93 pKa = 4.01RR94 pKa = 11.84PFLIQVVPDD103 pKa = 3.55SRR105 pKa = 11.84FACYY109 pKa = 10.61VFTEE113 pKa = 4.42YY114 pKa = 10.93VQQNYY119 pKa = 10.17SKK121 pKa = 10.8FDD123 pKa = 3.62VEE125 pKa = 4.47SSSMVSPATVVGYY138 pKa = 10.76LMYY141 pKa = 10.3CLHH144 pKa = 6.93AFIFLTDD151 pKa = 4.0VYY153 pKa = 11.09SSSTMSAYY161 pKa = 10.32AEE163 pKa = 4.73EE164 pKa = 4.81IDD166 pKa = 4.33ASHH169 pKa = 7.23IMRR172 pKa = 11.84KK173 pKa = 10.11LIDD176 pKa = 3.6IFSNCLVPDD185 pKa = 3.71IVFSVLDD192 pKa = 3.71ALHH195 pKa = 6.04PHH197 pKa = 6.57KK198 pKa = 10.79LDD200 pKa = 3.11VRR202 pKa = 11.84TTVSLFPSYY211 pKa = 11.16GSVLFEE217 pKa = 3.96YY218 pKa = 10.08DD219 pKa = 3.67APRR222 pKa = 11.84LIPPSIFLLAHH233 pKa = 5.89NQLLTQTKK241 pKa = 9.39VPNSYY246 pKa = 10.65KK247 pKa = 10.14SWLSQTCVTYY257 pKa = 8.92NTEE260 pKa = 4.21DD261 pKa = 3.57YY262 pKa = 10.77RR263 pKa = 11.84VGNIIGGIYY272 pKa = 7.73QTISGSTTEE281 pKa = 4.06TFQYY285 pKa = 8.85KK286 pKa = 8.21TWLLRR291 pKa = 11.84SLSRR295 pKa = 11.84LADD298 pKa = 3.5SATHH302 pKa = 5.62RR303 pKa = 11.84THH305 pKa = 7.63LKK307 pKa = 9.87RR308 pKa = 11.84NAVTEE313 pKa = 4.1IQFDD317 pKa = 3.99PPSFTDD323 pKa = 4.01ADD325 pKa = 4.26FNPYY329 pKa = 9.58TYY331 pKa = 11.5LLMLSPSSRR340 pKa = 11.84TTTTSFLTALSSSCQKK356 pKa = 10.49YY357 pKa = 10.81LNATRR362 pKa = 11.84SLSSVLATRR371 pKa = 11.84SGSITRR377 pKa = 11.84HH378 pKa = 5.44MIFDD382 pKa = 4.25LTAPTANYY390 pKa = 7.58STLIDD395 pKa = 4.09LDD397 pKa = 3.85EE398 pKa = 5.33SSVLKK403 pKa = 10.41PGNFANFCKK412 pKa = 10.4AVDD415 pKa = 4.36FCPPQISNPPKK426 pKa = 10.52KK427 pKa = 9.55LTLPYY432 pKa = 9.57PPGVEE437 pKa = 4.03AHH439 pKa = 6.7PSLYY443 pKa = 10.63LVTEE447 pKa = 4.48KK448 pKa = 10.75EE449 pKa = 4.35SEE451 pKa = 4.41SPLKK455 pKa = 9.99PLRR458 pKa = 11.84PSYY461 pKa = 9.96EE462 pKa = 3.92DD463 pKa = 3.29HH464 pKa = 7.77VEE466 pKa = 4.04GQILLFDD473 pKa = 5.31PYY475 pKa = 11.24DD476 pKa = 4.51DD477 pKa = 4.37EE478 pKa = 5.77PSAHH482 pKa = 6.81FSTLISGKK490 pKa = 10.35LIEE493 pKa = 4.96NGNVDD498 pKa = 4.78GITIFLPSPSSSISAVNSRR517 pKa = 11.84NLQGAIPLCRR527 pKa = 11.84IQATFAGFPFQQYY540 pKa = 8.92PRR542 pKa = 11.84SHH544 pKa = 6.49APSSRR549 pKa = 11.84MFTMSILYY557 pKa = 9.8DD558 pKa = 3.71AANIWLPYY566 pKa = 8.77FNRR569 pKa = 11.84VMYY572 pKa = 10.23SALPILPYY580 pKa = 9.08TANRR584 pKa = 11.84NADD587 pKa = 3.53GLVPSTNLVVTSSKK601 pKa = 9.71TVSTPSFSPDD611 pKa = 2.71VSLWSSYY618 pKa = 10.07RR619 pKa = 11.84YY620 pKa = 9.66QPTNEE625 pKa = 4.22RR626 pKa = 11.84PTPQNVYY633 pKa = 9.49MYY635 pKa = 8.81ATLEE639 pKa = 4.09PFFGSRR645 pKa = 11.84SSYY648 pKa = 9.03LQSYY652 pKa = 7.65QLQVLLPISS661 pKa = 3.62
MM1 pKa = 7.15SHH3 pKa = 7.84DD4 pKa = 3.89ARR6 pKa = 11.84DD7 pKa = 3.75LQSEE11 pKa = 4.18ARR13 pKa = 11.84QEE15 pKa = 3.93VRR17 pKa = 11.84PSGLQPDD24 pKa = 3.99AHH26 pKa = 6.53SHH28 pKa = 5.66EE29 pKa = 5.37PIGLNNSIARR39 pKa = 11.84TAPSSSAGPGLTAPHH54 pKa = 6.94AGTRR58 pKa = 11.84SLGLPCKK65 pKa = 10.65GEE67 pKa = 4.03ASHH70 pKa = 7.34SSEE73 pKa = 4.0ASSLARR79 pKa = 11.84DD80 pKa = 3.02IDD82 pKa = 3.69LGRR85 pKa = 11.84NCSRR89 pKa = 11.84SSGEE93 pKa = 4.01RR94 pKa = 11.84PFLIQVVPDD103 pKa = 3.55SRR105 pKa = 11.84FACYY109 pKa = 10.61VFTEE113 pKa = 4.42YY114 pKa = 10.93VQQNYY119 pKa = 10.17SKK121 pKa = 10.8FDD123 pKa = 3.62VEE125 pKa = 4.47SSSMVSPATVVGYY138 pKa = 10.76LMYY141 pKa = 10.3CLHH144 pKa = 6.93AFIFLTDD151 pKa = 4.0VYY153 pKa = 11.09SSSTMSAYY161 pKa = 10.32AEE163 pKa = 4.73EE164 pKa = 4.81IDD166 pKa = 4.33ASHH169 pKa = 7.23IMRR172 pKa = 11.84KK173 pKa = 10.11LIDD176 pKa = 3.6IFSNCLVPDD185 pKa = 3.71IVFSVLDD192 pKa = 3.71ALHH195 pKa = 6.04PHH197 pKa = 6.57KK198 pKa = 10.79LDD200 pKa = 3.11VRR202 pKa = 11.84TTVSLFPSYY211 pKa = 11.16GSVLFEE217 pKa = 3.96YY218 pKa = 10.08DD219 pKa = 3.67APRR222 pKa = 11.84LIPPSIFLLAHH233 pKa = 5.89NQLLTQTKK241 pKa = 9.39VPNSYY246 pKa = 10.65KK247 pKa = 10.14SWLSQTCVTYY257 pKa = 8.92NTEE260 pKa = 4.21DD261 pKa = 3.57YY262 pKa = 10.77RR263 pKa = 11.84VGNIIGGIYY272 pKa = 7.73QTISGSTTEE281 pKa = 4.06TFQYY285 pKa = 8.85KK286 pKa = 8.21TWLLRR291 pKa = 11.84SLSRR295 pKa = 11.84LADD298 pKa = 3.5SATHH302 pKa = 5.62RR303 pKa = 11.84THH305 pKa = 7.63LKK307 pKa = 9.87RR308 pKa = 11.84NAVTEE313 pKa = 4.1IQFDD317 pKa = 3.99PPSFTDD323 pKa = 4.01ADD325 pKa = 4.26FNPYY329 pKa = 9.58TYY331 pKa = 11.5LLMLSPSSRR340 pKa = 11.84TTTTSFLTALSSSCQKK356 pKa = 10.49YY357 pKa = 10.81LNATRR362 pKa = 11.84SLSSVLATRR371 pKa = 11.84SGSITRR377 pKa = 11.84HH378 pKa = 5.44MIFDD382 pKa = 4.25LTAPTANYY390 pKa = 7.58STLIDD395 pKa = 4.09LDD397 pKa = 3.85EE398 pKa = 5.33SSVLKK403 pKa = 10.41PGNFANFCKK412 pKa = 10.4AVDD415 pKa = 4.36FCPPQISNPPKK426 pKa = 10.52KK427 pKa = 9.55LTLPYY432 pKa = 9.57PPGVEE437 pKa = 4.03AHH439 pKa = 6.7PSLYY443 pKa = 10.63LVTEE447 pKa = 4.48KK448 pKa = 10.75EE449 pKa = 4.35SEE451 pKa = 4.41SPLKK455 pKa = 9.99PLRR458 pKa = 11.84PSYY461 pKa = 9.96EE462 pKa = 3.92DD463 pKa = 3.29HH464 pKa = 7.77VEE466 pKa = 4.04GQILLFDD473 pKa = 5.31PYY475 pKa = 11.24DD476 pKa = 4.51DD477 pKa = 4.37EE478 pKa = 5.77PSAHH482 pKa = 6.81FSTLISGKK490 pKa = 10.35LIEE493 pKa = 4.96NGNVDD498 pKa = 4.78GITIFLPSPSSSISAVNSRR517 pKa = 11.84NLQGAIPLCRR527 pKa = 11.84IQATFAGFPFQQYY540 pKa = 8.92PRR542 pKa = 11.84SHH544 pKa = 6.49APSSRR549 pKa = 11.84MFTMSILYY557 pKa = 9.8DD558 pKa = 3.71AANIWLPYY566 pKa = 8.77FNRR569 pKa = 11.84VMYY572 pKa = 10.23SALPILPYY580 pKa = 9.08TANRR584 pKa = 11.84NADD587 pKa = 3.53GLVPSTNLVVTSSKK601 pKa = 9.71TVSTPSFSPDD611 pKa = 2.71VSLWSSYY618 pKa = 10.07RR619 pKa = 11.84YY620 pKa = 9.66QPTNEE625 pKa = 4.22RR626 pKa = 11.84PTPQNVYY633 pKa = 9.49MYY635 pKa = 8.81ATLEE639 pKa = 4.09PFFGSRR645 pKa = 11.84SSYY648 pKa = 9.03LQSYY652 pKa = 7.65QLQVLLPISS661 pKa = 3.62
Molecular weight: 73.17 kDa
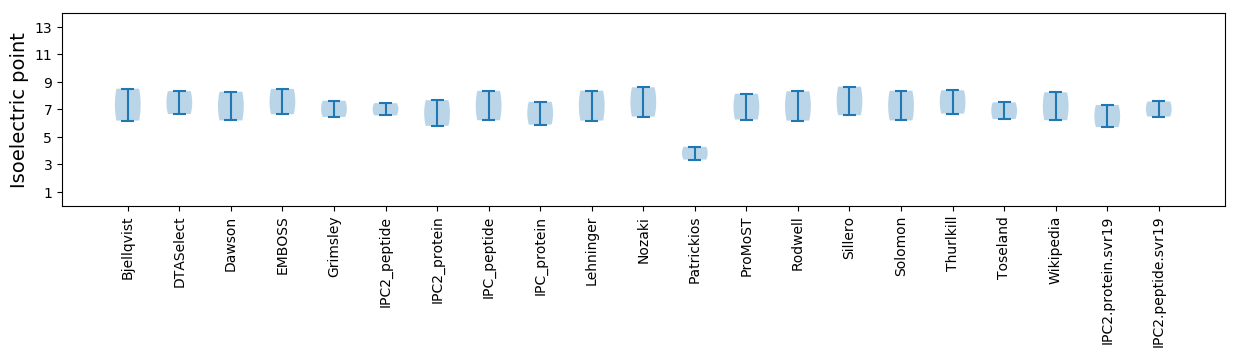
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6X9T6|Q6X9T6_9VIRU Putative RNA-dependent RNA polymerase OS=Ceratocystis polonica partitivirus OX=235434 GN=RDRP PE=4 SV=1
MM1 pKa = 7.9PSFSANPTYY10 pKa = 9.53QTLIDD15 pKa = 5.93DD16 pKa = 4.15IDD18 pKa = 4.71FDD20 pKa = 4.55VPIAHH25 pKa = 7.36PFSVINTDD33 pKa = 3.72LQPVDD38 pKa = 4.91DD39 pKa = 5.15EE40 pKa = 4.72EE41 pKa = 4.7TVHH44 pKa = 7.38DD45 pKa = 5.04LGSKK49 pKa = 10.41DD50 pKa = 3.34FEE52 pKa = 4.57FYY54 pKa = 10.89KK55 pKa = 10.79VVSDD59 pKa = 3.79NLPPTRR65 pKa = 11.84APSIGIEE72 pKa = 3.95SLPNIRR78 pKa = 11.84YY79 pKa = 8.9HH80 pKa = 5.68NHH82 pKa = 6.0SNDD85 pKa = 2.53HH86 pKa = 7.1RR87 pKa = 11.84YY88 pKa = 9.66RR89 pKa = 11.84DD90 pKa = 3.88QPPTGPPPMRR100 pKa = 11.84GVRR103 pKa = 11.84KK104 pKa = 9.91IINDD108 pKa = 3.62SFPQYY113 pKa = 10.94LPYY116 pKa = 10.56LKK118 pKa = 9.89EE119 pKa = 3.33WCRR122 pKa = 11.84PKK124 pKa = 10.42TSSDD128 pKa = 3.95AIFEE132 pKa = 4.84DD133 pKa = 5.28FNQPQIPSIPLSYY146 pKa = 10.39NRR148 pKa = 11.84KK149 pKa = 8.37QRR151 pKa = 11.84ILNLVNHH158 pKa = 6.29FMGVKK163 pKa = 9.81PYY165 pKa = 10.58DD166 pKa = 3.31IVHH169 pKa = 6.32FCDD172 pKa = 3.01TRR174 pKa = 11.84FYY176 pKa = 10.71PWDD179 pKa = 3.66LSKK182 pKa = 11.18KK183 pKa = 9.61ADD185 pKa = 3.61YY186 pKa = 10.16FHH188 pKa = 6.67NHH190 pKa = 3.66SNARR194 pKa = 11.84KK195 pKa = 8.35RR196 pKa = 11.84HH197 pKa = 5.02AQTSHH202 pKa = 6.64ASTATGPTKK211 pKa = 10.57KK212 pKa = 10.02SWFINAHH219 pKa = 6.13LFHH222 pKa = 7.45DD223 pKa = 4.68RR224 pKa = 11.84STVHH228 pKa = 6.61NIKK231 pKa = 10.7LYY233 pKa = 9.86GLPFKK238 pKa = 9.99PHH240 pKa = 6.52SYY242 pKa = 9.37EE243 pKa = 3.77ARR245 pKa = 11.84NKK247 pKa = 10.08ILLEE251 pKa = 3.93LWFKK255 pKa = 11.05KK256 pKa = 10.31IPTEE260 pKa = 3.77LLVRR264 pKa = 11.84SHH266 pKa = 7.07ISNPKK271 pKa = 7.83KK272 pKa = 10.73LKK274 pKa = 9.98VRR276 pKa = 11.84PVNNAPMIFLRR287 pKa = 11.84IEE289 pKa = 3.95CMLFYY294 pKa = 10.57PLLAQLRR301 pKa = 11.84KK302 pKa = 7.74QQCSIMYY309 pKa = 9.82GLEE312 pKa = 4.19TIRR315 pKa = 11.84GGMMEE320 pKa = 4.35IEE322 pKa = 4.23SLATRR327 pKa = 11.84FSNFMMIDD335 pKa = 3.05WSKK338 pKa = 11.11FDD340 pKa = 3.26QTVPFTLVDD349 pKa = 3.2MFYY352 pKa = 10.9QDD354 pKa = 4.79WIPTLILVDD363 pKa = 3.38SGYY366 pKa = 11.66AKK368 pKa = 9.51IHH370 pKa = 6.27NYY372 pKa = 9.35NDD374 pKa = 3.68HH375 pKa = 5.51VHH377 pKa = 6.44SFAAQARR384 pKa = 11.84KK385 pKa = 10.07LGVHH389 pKa = 6.7GDD391 pKa = 3.75SNLNEE396 pKa = 4.84APPEE400 pKa = 3.94AAVFANKK407 pKa = 9.76VEE409 pKa = 4.08NLLKK413 pKa = 10.72FINTWFKK420 pKa = 10.55EE421 pKa = 4.21MVYY424 pKa = 8.81ITPDD428 pKa = 2.69GFAYY432 pKa = 10.53SRR434 pKa = 11.84TFAGVPSGILCTQLIDD450 pKa = 3.71SFVNLVVLIDD460 pKa = 3.59SLFEE464 pKa = 4.18FGFHH468 pKa = 6.82EE469 pKa = 4.67SDD471 pKa = 3.13IKK473 pKa = 11.04SALILLMGDD482 pKa = 3.8DD483 pKa = 4.13NVVFAPDD490 pKa = 4.16KK491 pKa = 10.85LSQLHH496 pKa = 6.28SFFKK500 pKa = 10.61FLPDD504 pKa = 3.42YY505 pKa = 10.98AKK507 pKa = 10.6KK508 pKa = 9.68RR509 pKa = 11.84WNMKK513 pKa = 9.67VNVDD517 pKa = 3.29KK518 pKa = 11.43SIFTTLRR525 pKa = 11.84RR526 pKa = 11.84KK527 pKa = 9.82IEE529 pKa = 3.67ILGYY533 pKa = 7.61TNNYY537 pKa = 8.42GMPVRR542 pKa = 11.84SLSKK546 pKa = 10.81LIGQLAYY553 pKa = 9.75PEE555 pKa = 4.29RR556 pKa = 11.84HH557 pKa = 6.15VNDD560 pKa = 3.75SDD562 pKa = 3.22MCMRR566 pKa = 11.84AIGFAWCAAASDD578 pKa = 4.22STFHH582 pKa = 6.97DD583 pKa = 4.67FCRR586 pKa = 11.84KK587 pKa = 8.92VFIYY591 pKa = 10.29YY592 pKa = 8.45YY593 pKa = 11.0ARR595 pKa = 11.84VNVPIKK601 pKa = 10.89DD602 pKa = 3.85LVQSNASALPGMFFAYY618 pKa = 10.0RR619 pKa = 11.84DD620 pKa = 3.61VHH622 pKa = 5.18QHH624 pKa = 5.23IKK626 pKa = 10.48LDD628 pKa = 3.63HH629 pKa = 6.35FPSIEE634 pKa = 3.96EE635 pKa = 3.88VRR637 pKa = 11.84QVLSKK642 pKa = 10.21HH643 pKa = 5.82HH644 pKa = 6.94GYY646 pKa = 8.26LTEE649 pKa = 4.23EE650 pKa = 4.55PLWKK654 pKa = 10.22YY655 pKa = 11.4DD656 pKa = 3.65FFLHH660 pKa = 6.57PKK662 pKa = 8.8PP663 pKa = 4.15
MM1 pKa = 7.9PSFSANPTYY10 pKa = 9.53QTLIDD15 pKa = 5.93DD16 pKa = 4.15IDD18 pKa = 4.71FDD20 pKa = 4.55VPIAHH25 pKa = 7.36PFSVINTDD33 pKa = 3.72LQPVDD38 pKa = 4.91DD39 pKa = 5.15EE40 pKa = 4.72EE41 pKa = 4.7TVHH44 pKa = 7.38DD45 pKa = 5.04LGSKK49 pKa = 10.41DD50 pKa = 3.34FEE52 pKa = 4.57FYY54 pKa = 10.89KK55 pKa = 10.79VVSDD59 pKa = 3.79NLPPTRR65 pKa = 11.84APSIGIEE72 pKa = 3.95SLPNIRR78 pKa = 11.84YY79 pKa = 8.9HH80 pKa = 5.68NHH82 pKa = 6.0SNDD85 pKa = 2.53HH86 pKa = 7.1RR87 pKa = 11.84YY88 pKa = 9.66RR89 pKa = 11.84DD90 pKa = 3.88QPPTGPPPMRR100 pKa = 11.84GVRR103 pKa = 11.84KK104 pKa = 9.91IINDD108 pKa = 3.62SFPQYY113 pKa = 10.94LPYY116 pKa = 10.56LKK118 pKa = 9.89EE119 pKa = 3.33WCRR122 pKa = 11.84PKK124 pKa = 10.42TSSDD128 pKa = 3.95AIFEE132 pKa = 4.84DD133 pKa = 5.28FNQPQIPSIPLSYY146 pKa = 10.39NRR148 pKa = 11.84KK149 pKa = 8.37QRR151 pKa = 11.84ILNLVNHH158 pKa = 6.29FMGVKK163 pKa = 9.81PYY165 pKa = 10.58DD166 pKa = 3.31IVHH169 pKa = 6.32FCDD172 pKa = 3.01TRR174 pKa = 11.84FYY176 pKa = 10.71PWDD179 pKa = 3.66LSKK182 pKa = 11.18KK183 pKa = 9.61ADD185 pKa = 3.61YY186 pKa = 10.16FHH188 pKa = 6.67NHH190 pKa = 3.66SNARR194 pKa = 11.84KK195 pKa = 8.35RR196 pKa = 11.84HH197 pKa = 5.02AQTSHH202 pKa = 6.64ASTATGPTKK211 pKa = 10.57KK212 pKa = 10.02SWFINAHH219 pKa = 6.13LFHH222 pKa = 7.45DD223 pKa = 4.68RR224 pKa = 11.84STVHH228 pKa = 6.61NIKK231 pKa = 10.7LYY233 pKa = 9.86GLPFKK238 pKa = 9.99PHH240 pKa = 6.52SYY242 pKa = 9.37EE243 pKa = 3.77ARR245 pKa = 11.84NKK247 pKa = 10.08ILLEE251 pKa = 3.93LWFKK255 pKa = 11.05KK256 pKa = 10.31IPTEE260 pKa = 3.77LLVRR264 pKa = 11.84SHH266 pKa = 7.07ISNPKK271 pKa = 7.83KK272 pKa = 10.73LKK274 pKa = 9.98VRR276 pKa = 11.84PVNNAPMIFLRR287 pKa = 11.84IEE289 pKa = 3.95CMLFYY294 pKa = 10.57PLLAQLRR301 pKa = 11.84KK302 pKa = 7.74QQCSIMYY309 pKa = 9.82GLEE312 pKa = 4.19TIRR315 pKa = 11.84GGMMEE320 pKa = 4.35IEE322 pKa = 4.23SLATRR327 pKa = 11.84FSNFMMIDD335 pKa = 3.05WSKK338 pKa = 11.11FDD340 pKa = 3.26QTVPFTLVDD349 pKa = 3.2MFYY352 pKa = 10.9QDD354 pKa = 4.79WIPTLILVDD363 pKa = 3.38SGYY366 pKa = 11.66AKK368 pKa = 9.51IHH370 pKa = 6.27NYY372 pKa = 9.35NDD374 pKa = 3.68HH375 pKa = 5.51VHH377 pKa = 6.44SFAAQARR384 pKa = 11.84KK385 pKa = 10.07LGVHH389 pKa = 6.7GDD391 pKa = 3.75SNLNEE396 pKa = 4.84APPEE400 pKa = 3.94AAVFANKK407 pKa = 9.76VEE409 pKa = 4.08NLLKK413 pKa = 10.72FINTWFKK420 pKa = 10.55EE421 pKa = 4.21MVYY424 pKa = 8.81ITPDD428 pKa = 2.69GFAYY432 pKa = 10.53SRR434 pKa = 11.84TFAGVPSGILCTQLIDD450 pKa = 3.71SFVNLVVLIDD460 pKa = 3.59SLFEE464 pKa = 4.18FGFHH468 pKa = 6.82EE469 pKa = 4.67SDD471 pKa = 3.13IKK473 pKa = 11.04SALILLMGDD482 pKa = 3.8DD483 pKa = 4.13NVVFAPDD490 pKa = 4.16KK491 pKa = 10.85LSQLHH496 pKa = 6.28SFFKK500 pKa = 10.61FLPDD504 pKa = 3.42YY505 pKa = 10.98AKK507 pKa = 10.6KK508 pKa = 9.68RR509 pKa = 11.84WNMKK513 pKa = 9.67VNVDD517 pKa = 3.29KK518 pKa = 11.43SIFTTLRR525 pKa = 11.84RR526 pKa = 11.84KK527 pKa = 9.82IEE529 pKa = 3.67ILGYY533 pKa = 7.61TNNYY537 pKa = 8.42GMPVRR542 pKa = 11.84SLSKK546 pKa = 10.81LIGQLAYY553 pKa = 9.75PEE555 pKa = 4.29RR556 pKa = 11.84HH557 pKa = 6.15VNDD560 pKa = 3.75SDD562 pKa = 3.22MCMRR566 pKa = 11.84AIGFAWCAAASDD578 pKa = 4.22STFHH582 pKa = 6.97DD583 pKa = 4.67FCRR586 pKa = 11.84KK587 pKa = 8.92VFIYY591 pKa = 10.29YY592 pKa = 8.45YY593 pKa = 11.0ARR595 pKa = 11.84VNVPIKK601 pKa = 10.89DD602 pKa = 3.85LVQSNASALPGMFFAYY618 pKa = 10.0RR619 pKa = 11.84DD620 pKa = 3.61VHH622 pKa = 5.18QHH624 pKa = 5.23IKK626 pKa = 10.48LDD628 pKa = 3.63HH629 pKa = 6.35FPSIEE634 pKa = 3.96EE635 pKa = 3.88VRR637 pKa = 11.84QVLSKK642 pKa = 10.21HH643 pKa = 5.82HH644 pKa = 6.94GYY646 pKa = 8.26LTEE649 pKa = 4.23EE650 pKa = 4.55PLWKK654 pKa = 10.22YY655 pKa = 11.4DD656 pKa = 3.65FFLHH660 pKa = 6.57PKK662 pKa = 8.8PP663 pKa = 4.15
Molecular weight: 76.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1324 |
661 |
663 |
662.0 |
75.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.118 ± 0.4 | 1.36 ± 0.114 |
5.665 ± 0.724 | 3.776 ± 0.004 |
5.891 ± 0.892 | 3.776 ± 0.117 |
3.55 ± 0.726 | 5.816 ± 0.611 |
4.456 ± 1.399 | 9.366 ± 0.572 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.19 ± 0.391 | 4.758 ± 0.5 |
7.477 ± 0.402 | 3.323 ± 0.228 |
4.834 ± 0.005 | 10.423 ± 2.483 |
5.816 ± 1.186 | 5.665 ± 0.162 |
1.057 ± 0.336 | 4.683 ± 0.23 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
