
Gloeobacter violaceus (strain ATCC 29082 / PCC 7421)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Gloeobacteria; Gloeobacterales; Gloeobacteraceae; Gloeobacter; Gloeobacter violaceus
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
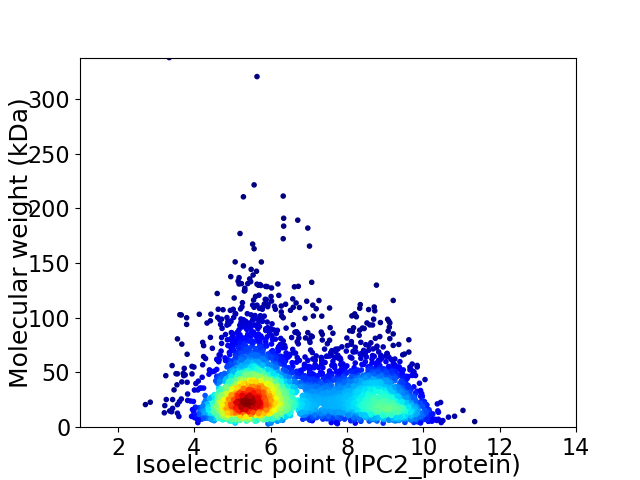
Virtual 2D-PAGE plot for 4406 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q7NK40|Q7NK40_GLOVI Gll1640 protein OS=Gloeobacter violaceus (strain ATCC 29082 / PCC 7421) OX=251221 GN=gll1640 PE=4 SV=1
MM1 pKa = 7.21EE2 pKa = 5.26EE3 pKa = 5.22KK4 pKa = 10.95YY5 pKa = 10.65FLQNEE10 pKa = 4.41SDD12 pKa = 3.75GSGDD16 pKa = 3.04ATYY19 pKa = 10.46IGTLGEE25 pKa = 4.22EE26 pKa = 4.75GAQVNAFSQNGEE38 pKa = 3.85YY39 pKa = 10.41SYY41 pKa = 11.85GSDD44 pKa = 3.58GNQNVLWDD52 pKa = 3.86QDD54 pKa = 3.07SDD56 pKa = 4.48YY57 pKa = 11.7YY58 pKa = 11.29SVQNLYY64 pKa = 10.82ASEE67 pKa = 4.97DD68 pKa = 3.85GSSLDD73 pKa = 4.04SGTGSTNFGNFEE85 pKa = 4.17NAWISGSASLSDD97 pKa = 3.61NEE99 pKa = 4.66QDD101 pKa = 3.1IDD103 pKa = 4.2TNSDD107 pKa = 3.47VYY109 pKa = 11.51DD110 pKa = 4.65SIAQGVSGLDD120 pKa = 3.33FDD122 pKa = 5.61FDD124 pKa = 4.21SFSDD128 pKa = 3.56ANGTVFATNADD139 pKa = 4.26SPNSNYY145 pKa = 10.04PGADD149 pKa = 2.64TYY151 pKa = 11.89GAMNDD156 pKa = 3.31DD157 pKa = 5.15GYY159 pKa = 11.61RR160 pKa = 11.84LLPGASAVTDD170 pKa = 3.46SAIPQFEE177 pKa = 4.09QAATDD182 pKa = 3.33QDD184 pKa = 4.15LPLAADD190 pKa = 3.62EE191 pKa = 4.79LLVVDD196 pKa = 4.78AQIEE200 pKa = 4.42VEE202 pKa = 4.01NDD204 pKa = 2.32IHH206 pKa = 7.41ALLVSSGEE214 pKa = 3.9QASIAFSLPQGANADD229 pKa = 3.83FMNDD233 pKa = 3.19SSVMAIAKK241 pKa = 9.02WDD243 pKa = 3.76EE244 pKa = 4.29LGGSSNGQTDD254 pKa = 2.87SWNADD259 pKa = 3.74EE260 pKa = 5.47LNVIAGKK267 pKa = 10.49LPAGPNGYY275 pKa = 9.13PDD277 pKa = 5.51DD278 pKa = 3.89KK279 pKa = 10.7TCRR282 pKa = 11.84LLHH285 pKa = 6.12EE286 pKa = 5.41LEE288 pKa = 4.2QKK290 pKa = 11.06YY291 pKa = 10.5PDD293 pKa = 3.63TPQQKK298 pKa = 9.2RR299 pKa = 11.84RR300 pKa = 11.84EE301 pKa = 4.07EE302 pKa = 4.02EE303 pKa = 3.78RR304 pKa = 11.84RR305 pKa = 11.84SMEE308 pKa = 3.88QITYY312 pKa = 9.34PGAGGMLGTIVGSGLTGAAAGSAAGSLGFGIGGLVGGAIGGLAGTIAGEE361 pKa = 4.01QASQDD366 pKa = 3.91KK367 pKa = 10.83VWVEE371 pKa = 3.9DD372 pKa = 3.98AYY374 pKa = 10.21KK375 pKa = 10.35HH376 pKa = 6.48CEE378 pKa = 4.06EE379 pKa = 5.23QGTQAPWTRR388 pKa = 11.84PEE390 pKa = 3.89PPRR393 pKa = 5.45
MM1 pKa = 7.21EE2 pKa = 5.26EE3 pKa = 5.22KK4 pKa = 10.95YY5 pKa = 10.65FLQNEE10 pKa = 4.41SDD12 pKa = 3.75GSGDD16 pKa = 3.04ATYY19 pKa = 10.46IGTLGEE25 pKa = 4.22EE26 pKa = 4.75GAQVNAFSQNGEE38 pKa = 3.85YY39 pKa = 10.41SYY41 pKa = 11.85GSDD44 pKa = 3.58GNQNVLWDD52 pKa = 3.86QDD54 pKa = 3.07SDD56 pKa = 4.48YY57 pKa = 11.7YY58 pKa = 11.29SVQNLYY64 pKa = 10.82ASEE67 pKa = 4.97DD68 pKa = 3.85GSSLDD73 pKa = 4.04SGTGSTNFGNFEE85 pKa = 4.17NAWISGSASLSDD97 pKa = 3.61NEE99 pKa = 4.66QDD101 pKa = 3.1IDD103 pKa = 4.2TNSDD107 pKa = 3.47VYY109 pKa = 11.51DD110 pKa = 4.65SIAQGVSGLDD120 pKa = 3.33FDD122 pKa = 5.61FDD124 pKa = 4.21SFSDD128 pKa = 3.56ANGTVFATNADD139 pKa = 4.26SPNSNYY145 pKa = 10.04PGADD149 pKa = 2.64TYY151 pKa = 11.89GAMNDD156 pKa = 3.31DD157 pKa = 5.15GYY159 pKa = 11.61RR160 pKa = 11.84LLPGASAVTDD170 pKa = 3.46SAIPQFEE177 pKa = 4.09QAATDD182 pKa = 3.33QDD184 pKa = 4.15LPLAADD190 pKa = 3.62EE191 pKa = 4.79LLVVDD196 pKa = 4.78AQIEE200 pKa = 4.42VEE202 pKa = 4.01NDD204 pKa = 2.32IHH206 pKa = 7.41ALLVSSGEE214 pKa = 3.9QASIAFSLPQGANADD229 pKa = 3.83FMNDD233 pKa = 3.19SSVMAIAKK241 pKa = 9.02WDD243 pKa = 3.76EE244 pKa = 4.29LGGSSNGQTDD254 pKa = 2.87SWNADD259 pKa = 3.74EE260 pKa = 5.47LNVIAGKK267 pKa = 10.49LPAGPNGYY275 pKa = 9.13PDD277 pKa = 5.51DD278 pKa = 3.89KK279 pKa = 10.7TCRR282 pKa = 11.84LLHH285 pKa = 6.12EE286 pKa = 5.41LEE288 pKa = 4.2QKK290 pKa = 11.06YY291 pKa = 10.5PDD293 pKa = 3.63TPQQKK298 pKa = 9.2RR299 pKa = 11.84RR300 pKa = 11.84EE301 pKa = 4.07EE302 pKa = 4.02EE303 pKa = 3.78RR304 pKa = 11.84RR305 pKa = 11.84SMEE308 pKa = 3.88QITYY312 pKa = 9.34PGAGGMLGTIVGSGLTGAAAGSAAGSLGFGIGGLVGGAIGGLAGTIAGEE361 pKa = 4.01QASQDD366 pKa = 3.91KK367 pKa = 10.83VWVEE371 pKa = 3.9DD372 pKa = 3.98AYY374 pKa = 10.21KK375 pKa = 10.35HH376 pKa = 6.48CEE378 pKa = 4.06EE379 pKa = 5.23QGTQAPWTRR388 pKa = 11.84PEE390 pKa = 3.89PPRR393 pKa = 5.45
Molecular weight: 41.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q7NKK7|Q7NKK7_GLOVI Gll1470 protein OS=Gloeobacter violaceus (strain ATCC 29082 / PCC 7421) OX=251221 GN=gll1470 PE=4 SV=1
MM1 pKa = 7.39KK2 pKa = 8.95RR3 pKa = 11.84TLGGTTRR10 pKa = 11.84KK11 pKa = 9.29RR12 pKa = 11.84QKK14 pKa = 9.42TSGFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TASGRR28 pKa = 11.84RR29 pKa = 11.84VLSARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84HH40 pKa = 4.58RR41 pKa = 11.84LAVV44 pKa = 3.46
MM1 pKa = 7.39KK2 pKa = 8.95RR3 pKa = 11.84TLGGTTRR10 pKa = 11.84KK11 pKa = 9.29RR12 pKa = 11.84QKK14 pKa = 9.42TSGFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TASGRR28 pKa = 11.84RR29 pKa = 11.84VLSARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84HH40 pKa = 4.58RR41 pKa = 11.84LAVV44 pKa = 3.46
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1377677 |
28 |
3277 |
312.7 |
34.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.172 ± 0.046 | 1.024 ± 0.015 |
4.941 ± 0.029 | 6.146 ± 0.047 |
3.734 ± 0.025 | 8.331 ± 0.04 |
1.87 ± 0.018 | 4.433 ± 0.027 |
2.889 ± 0.033 | 11.419 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.733 ± 0.015 | 2.71 ± 0.029 |
5.654 ± 0.04 | 4.182 ± 0.028 |
7.41 ± 0.044 | 5.485 ± 0.028 |
5.178 ± 0.028 | 7.548 ± 0.031 |
1.503 ± 0.019 | 2.638 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
