
Hubei toti-like virus 16
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
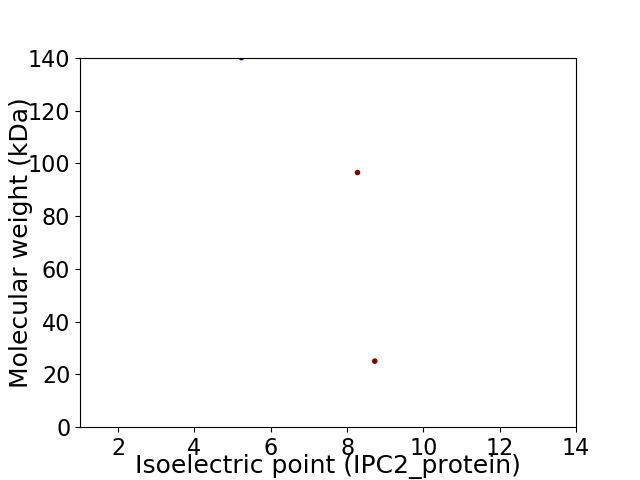
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFH4|A0A1L3KFH4_9VIRU Uncharacterized protein OS=Hubei toti-like virus 16 OX=1923304 PE=4 SV=1
MM1 pKa = 7.63AFLAPLVIPALQALGFGGLTALGSAAVKK29 pKa = 10.29KK30 pKa = 10.74VIGDD34 pKa = 4.19AVPEE38 pKa = 4.19DD39 pKa = 3.67QQIALLTSTIGGQMDD54 pKa = 4.1PSSTQSLSPQLQSGLMEE71 pKa = 5.39LKK73 pKa = 9.54TAMGAAPTLFFSQEE87 pKa = 3.44ALAPRR92 pKa = 11.84SAQGFLNQPLVSSHH106 pKa = 6.18SDD108 pKa = 3.3QRR110 pKa = 11.84AHH112 pKa = 6.64SALSQTLSDD121 pKa = 3.99AAALSQLRR129 pKa = 11.84GMQLRR134 pKa = 11.84LQSIDD139 pKa = 3.25PMATTRR145 pKa = 11.84DD146 pKa = 3.57PGVAAFPGSTPSTSLRR162 pKa = 11.84EE163 pKa = 4.18TRR165 pKa = 11.84DD166 pKa = 3.37PMQGQQLPGTSPASWLGLQQVSPPTFQNQLPLGMSPDD203 pKa = 3.59HH204 pKa = 6.18QQGQTTSSTLSMIPGTNLLVAPQPINKK231 pKa = 9.12LQDD234 pKa = 3.34NQAVLTEE241 pKa = 4.43RR242 pKa = 11.84PPAAGTQNPVAQGISGVGGSPGLGNIVRR270 pKa = 11.84GAGTEE275 pKa = 3.62IFQGVKK281 pKa = 9.93EE282 pKa = 4.36AGQAAASQAVSQISGTATDD301 pKa = 4.17LAKK304 pKa = 10.74KK305 pKa = 10.31GLTKK309 pKa = 10.72AGGYY313 pKa = 9.56LGSLISSGLTSIAKK327 pKa = 9.9KK328 pKa = 10.29IIGDD332 pKa = 3.86APPGFEE338 pKa = 5.0GFDD341 pKa = 3.31ARR343 pKa = 11.84KK344 pKa = 10.03GGFIGPTLDD353 pKa = 4.6DD354 pKa = 3.79VFTGGFNKK362 pKa = 10.24YY363 pKa = 9.78GRR365 pKa = 11.84EE366 pKa = 3.87TPMALASNVDD376 pKa = 3.8LNKK379 pKa = 10.06IVGDD383 pKa = 4.23APPGEE388 pKa = 4.36QGEE391 pKa = 4.71VMTDD395 pKa = 3.12AQAAAATTNLTNSVKK410 pKa = 10.13FNMMSRR416 pKa = 11.84IILNEE421 pKa = 3.53QTLTQSSSLPRR432 pKa = 11.84GMSEE436 pKa = 3.79LQLNNFFARR445 pKa = 11.84DD446 pKa = 3.69LEE448 pKa = 4.63SLVGTYY454 pKa = 9.49KK455 pKa = 10.44QSEE458 pKa = 4.35VQALFGFVGMTCQYY472 pKa = 10.24NRR474 pKa = 11.84WPAAVYY480 pKa = 9.33TPEE483 pKa = 4.11TVEE486 pKa = 4.5SKK488 pKa = 9.36GTPKK492 pKa = 10.56EE493 pKa = 4.03KK494 pKa = 10.55VIPAHH499 pKa = 6.16NGVGTFSIDD508 pKa = 3.06ALYY511 pKa = 8.67CTGVALEE518 pKa = 4.33TARR521 pKa = 11.84VAPPCTPNVTLSQLGPLLAGAALGEE546 pKa = 4.36YY547 pKa = 10.21SSQLRR552 pKa = 11.84VKK554 pKa = 9.7TFSTDD559 pKa = 2.49MDD561 pKa = 3.97LMIRR565 pKa = 11.84ASAEE569 pKa = 3.67ALNNIQSQQGYY580 pKa = 8.11SFCLPLTKK588 pKa = 10.48AIGYY592 pKa = 7.76VLQCIQLFGSEE603 pKa = 4.28FNEE606 pKa = 4.57LVADD610 pKa = 4.04MYY612 pKa = 10.94FRR614 pKa = 11.84PTEE617 pKa = 4.2KK618 pKa = 10.44GQHH621 pKa = 5.94HH622 pKa = 7.13AFQRR626 pKa = 11.84AFPLEE631 pKa = 4.14TLAGTEE637 pKa = 3.83AWEE640 pKa = 4.38GDD642 pKa = 3.34DD643 pKa = 3.39WQGAKK648 pKa = 10.03FAVVCEE654 pKa = 4.0SDD656 pKa = 4.15FIKK659 pKa = 9.68VTCGIISGANWGEE672 pKa = 3.84FQPAEE677 pKa = 4.05WGHH680 pKa = 5.55NCAVVFIPNNDD691 pKa = 3.34KK692 pKa = 11.33SNQLMNAVRR701 pKa = 11.84MISQMAYY708 pKa = 8.96PAKK711 pKa = 10.3LLYY714 pKa = 10.02IHH716 pKa = 6.86GKK718 pKa = 6.42WAYY721 pKa = 9.53FSRR724 pKa = 11.84EE725 pKa = 4.05GEE727 pKa = 4.08LTTIGAEE734 pKa = 3.69VHH736 pKa = 5.49AQKK739 pKa = 10.83KK740 pKa = 10.28AGLTRR745 pKa = 11.84IPGPYY750 pKa = 9.31KK751 pKa = 9.51YY752 pKa = 10.89ALFVVMGARR761 pKa = 11.84EE762 pKa = 3.86QSSAGIIIGDD772 pKa = 3.59GSGQANHH779 pKa = 6.23QIEE782 pKa = 4.96VNAIDD787 pKa = 4.5HH788 pKa = 6.6NPTTPYY794 pKa = 10.72VCDD797 pKa = 3.7AGINGFSAARR807 pKa = 11.84NGAVTDD813 pKa = 3.77AAVPMWMAFIKK824 pKa = 9.76EE825 pKa = 4.01WEE827 pKa = 4.34VIYY830 pKa = 10.99GNASDD835 pKa = 4.63RR836 pKa = 11.84SSAMRR841 pKa = 11.84FWADD845 pKa = 3.1NSILFGNFFLGKK857 pKa = 9.84NPLDD861 pKa = 3.8IGDD864 pKa = 3.86LQGFSFLSWADD875 pKa = 3.75PVAHH879 pKa = 7.07PPLPSCLAEE888 pKa = 4.09YY889 pKa = 10.37TDD891 pKa = 6.23LPVSWSGSCKK901 pKa = 9.93QVTPQVARR909 pKa = 11.84YY910 pKa = 9.35LMSSMKK916 pKa = 8.92TASYY920 pKa = 9.82MPVASITPDD929 pKa = 3.41YY930 pKa = 10.96ASQRR934 pKa = 11.84PWTWGYY940 pKa = 11.31GNPPDD945 pKa = 4.33GCRR948 pKa = 11.84LPYY951 pKa = 9.67EE952 pKa = 4.28DD953 pKa = 4.92VMISFLVNKK962 pKa = 9.9KK963 pKa = 8.93WISPNEE969 pKa = 3.97EE970 pKa = 3.98YY971 pKa = 10.27PAVSLDD977 pKa = 3.21DD978 pKa = 3.9AARR981 pKa = 11.84LGHH984 pKa = 7.22TITAMANIMAAAADD998 pKa = 3.57ILVQEE1003 pKa = 4.85RR1004 pKa = 11.84DD1005 pKa = 3.4FTLPEE1010 pKa = 4.32VIFPPLVLQRR1020 pKa = 11.84VPMAMMRR1027 pKa = 11.84QKK1029 pKa = 11.0NVLWPALEE1037 pKa = 4.02EE1038 pKa = 4.11LLTNGISYY1046 pKa = 8.24PQYY1049 pKa = 9.59WNNRR1053 pKa = 11.84DD1054 pKa = 3.43VNMHH1058 pKa = 6.15CWEE1061 pKa = 3.92AMIYY1065 pKa = 9.68WEE1067 pKa = 4.67VAAQTIYY1074 pKa = 10.86PEE1076 pKa = 4.55TVSHH1080 pKa = 6.63GSAVTTSARR1089 pKa = 11.84IPVSVMGQWFAPLYY1103 pKa = 8.59PQYY1106 pKa = 10.78FQLRR1110 pKa = 11.84LNLQVMSPQRR1120 pKa = 11.84TYY1122 pKa = 11.1ISINNVARR1130 pKa = 11.84AMFKK1134 pKa = 10.57VEE1136 pKa = 4.09YY1137 pKa = 10.61AEE1139 pKa = 4.18EE1140 pKa = 4.34TTGLLGKK1147 pKa = 10.05SLSPVSWLMWRR1158 pKa = 11.84LSLGVIISDD1167 pKa = 3.37TVSANNGWIVQCVNNGGVFDD1187 pKa = 4.68YY1188 pKa = 9.88YY1189 pKa = 10.66IHH1191 pKa = 6.66MEE1193 pKa = 4.05QPDD1196 pKa = 3.4ASRR1199 pKa = 11.84NQVMCRR1205 pKa = 11.84EE1206 pKa = 4.09YY1207 pKa = 10.77GAGNVTVPPTTKK1219 pKa = 9.87ISLPTGEE1226 pKa = 5.35FILKK1230 pKa = 10.3RR1231 pKa = 11.84SIATGLQPFSLLMQGPRR1248 pKa = 11.84DD1249 pKa = 3.55AFIGVGTGSTAYY1261 pKa = 10.88QMFTQTNTPQAIFVSEE1277 pKa = 4.26TWATIPDD1284 pKa = 4.01WLFSPAGLQSVRR1296 pKa = 11.84DD1297 pKa = 4.1SII1299 pKa = 4.59
MM1 pKa = 7.63AFLAPLVIPALQALGFGGLTALGSAAVKK29 pKa = 10.29KK30 pKa = 10.74VIGDD34 pKa = 4.19AVPEE38 pKa = 4.19DD39 pKa = 3.67QQIALLTSTIGGQMDD54 pKa = 4.1PSSTQSLSPQLQSGLMEE71 pKa = 5.39LKK73 pKa = 9.54TAMGAAPTLFFSQEE87 pKa = 3.44ALAPRR92 pKa = 11.84SAQGFLNQPLVSSHH106 pKa = 6.18SDD108 pKa = 3.3QRR110 pKa = 11.84AHH112 pKa = 6.64SALSQTLSDD121 pKa = 3.99AAALSQLRR129 pKa = 11.84GMQLRR134 pKa = 11.84LQSIDD139 pKa = 3.25PMATTRR145 pKa = 11.84DD146 pKa = 3.57PGVAAFPGSTPSTSLRR162 pKa = 11.84EE163 pKa = 4.18TRR165 pKa = 11.84DD166 pKa = 3.37PMQGQQLPGTSPASWLGLQQVSPPTFQNQLPLGMSPDD203 pKa = 3.59HH204 pKa = 6.18QQGQTTSSTLSMIPGTNLLVAPQPINKK231 pKa = 9.12LQDD234 pKa = 3.34NQAVLTEE241 pKa = 4.43RR242 pKa = 11.84PPAAGTQNPVAQGISGVGGSPGLGNIVRR270 pKa = 11.84GAGTEE275 pKa = 3.62IFQGVKK281 pKa = 9.93EE282 pKa = 4.36AGQAAASQAVSQISGTATDD301 pKa = 4.17LAKK304 pKa = 10.74KK305 pKa = 10.31GLTKK309 pKa = 10.72AGGYY313 pKa = 9.56LGSLISSGLTSIAKK327 pKa = 9.9KK328 pKa = 10.29IIGDD332 pKa = 3.86APPGFEE338 pKa = 5.0GFDD341 pKa = 3.31ARR343 pKa = 11.84KK344 pKa = 10.03GGFIGPTLDD353 pKa = 4.6DD354 pKa = 3.79VFTGGFNKK362 pKa = 10.24YY363 pKa = 9.78GRR365 pKa = 11.84EE366 pKa = 3.87TPMALASNVDD376 pKa = 3.8LNKK379 pKa = 10.06IVGDD383 pKa = 4.23APPGEE388 pKa = 4.36QGEE391 pKa = 4.71VMTDD395 pKa = 3.12AQAAAATTNLTNSVKK410 pKa = 10.13FNMMSRR416 pKa = 11.84IILNEE421 pKa = 3.53QTLTQSSSLPRR432 pKa = 11.84GMSEE436 pKa = 3.79LQLNNFFARR445 pKa = 11.84DD446 pKa = 3.69LEE448 pKa = 4.63SLVGTYY454 pKa = 9.49KK455 pKa = 10.44QSEE458 pKa = 4.35VQALFGFVGMTCQYY472 pKa = 10.24NRR474 pKa = 11.84WPAAVYY480 pKa = 9.33TPEE483 pKa = 4.11TVEE486 pKa = 4.5SKK488 pKa = 9.36GTPKK492 pKa = 10.56EE493 pKa = 4.03KK494 pKa = 10.55VIPAHH499 pKa = 6.16NGVGTFSIDD508 pKa = 3.06ALYY511 pKa = 8.67CTGVALEE518 pKa = 4.33TARR521 pKa = 11.84VAPPCTPNVTLSQLGPLLAGAALGEE546 pKa = 4.36YY547 pKa = 10.21SSQLRR552 pKa = 11.84VKK554 pKa = 9.7TFSTDD559 pKa = 2.49MDD561 pKa = 3.97LMIRR565 pKa = 11.84ASAEE569 pKa = 3.67ALNNIQSQQGYY580 pKa = 8.11SFCLPLTKK588 pKa = 10.48AIGYY592 pKa = 7.76VLQCIQLFGSEE603 pKa = 4.28FNEE606 pKa = 4.57LVADD610 pKa = 4.04MYY612 pKa = 10.94FRR614 pKa = 11.84PTEE617 pKa = 4.2KK618 pKa = 10.44GQHH621 pKa = 5.94HH622 pKa = 7.13AFQRR626 pKa = 11.84AFPLEE631 pKa = 4.14TLAGTEE637 pKa = 3.83AWEE640 pKa = 4.38GDD642 pKa = 3.34DD643 pKa = 3.39WQGAKK648 pKa = 10.03FAVVCEE654 pKa = 4.0SDD656 pKa = 4.15FIKK659 pKa = 9.68VTCGIISGANWGEE672 pKa = 3.84FQPAEE677 pKa = 4.05WGHH680 pKa = 5.55NCAVVFIPNNDD691 pKa = 3.34KK692 pKa = 11.33SNQLMNAVRR701 pKa = 11.84MISQMAYY708 pKa = 8.96PAKK711 pKa = 10.3LLYY714 pKa = 10.02IHH716 pKa = 6.86GKK718 pKa = 6.42WAYY721 pKa = 9.53FSRR724 pKa = 11.84EE725 pKa = 4.05GEE727 pKa = 4.08LTTIGAEE734 pKa = 3.69VHH736 pKa = 5.49AQKK739 pKa = 10.83KK740 pKa = 10.28AGLTRR745 pKa = 11.84IPGPYY750 pKa = 9.31KK751 pKa = 9.51YY752 pKa = 10.89ALFVVMGARR761 pKa = 11.84EE762 pKa = 3.86QSSAGIIIGDD772 pKa = 3.59GSGQANHH779 pKa = 6.23QIEE782 pKa = 4.96VNAIDD787 pKa = 4.5HH788 pKa = 6.6NPTTPYY794 pKa = 10.72VCDD797 pKa = 3.7AGINGFSAARR807 pKa = 11.84NGAVTDD813 pKa = 3.77AAVPMWMAFIKK824 pKa = 9.76EE825 pKa = 4.01WEE827 pKa = 4.34VIYY830 pKa = 10.99GNASDD835 pKa = 4.63RR836 pKa = 11.84SSAMRR841 pKa = 11.84FWADD845 pKa = 3.1NSILFGNFFLGKK857 pKa = 9.84NPLDD861 pKa = 3.8IGDD864 pKa = 3.86LQGFSFLSWADD875 pKa = 3.75PVAHH879 pKa = 7.07PPLPSCLAEE888 pKa = 4.09YY889 pKa = 10.37TDD891 pKa = 6.23LPVSWSGSCKK901 pKa = 9.93QVTPQVARR909 pKa = 11.84YY910 pKa = 9.35LMSSMKK916 pKa = 8.92TASYY920 pKa = 9.82MPVASITPDD929 pKa = 3.41YY930 pKa = 10.96ASQRR934 pKa = 11.84PWTWGYY940 pKa = 11.31GNPPDD945 pKa = 4.33GCRR948 pKa = 11.84LPYY951 pKa = 9.67EE952 pKa = 4.28DD953 pKa = 4.92VMISFLVNKK962 pKa = 9.9KK963 pKa = 8.93WISPNEE969 pKa = 3.97EE970 pKa = 3.98YY971 pKa = 10.27PAVSLDD977 pKa = 3.21DD978 pKa = 3.9AARR981 pKa = 11.84LGHH984 pKa = 7.22TITAMANIMAAAADD998 pKa = 3.57ILVQEE1003 pKa = 4.85RR1004 pKa = 11.84DD1005 pKa = 3.4FTLPEE1010 pKa = 4.32VIFPPLVLQRR1020 pKa = 11.84VPMAMMRR1027 pKa = 11.84QKK1029 pKa = 11.0NVLWPALEE1037 pKa = 4.02EE1038 pKa = 4.11LLTNGISYY1046 pKa = 8.24PQYY1049 pKa = 9.59WNNRR1053 pKa = 11.84DD1054 pKa = 3.43VNMHH1058 pKa = 6.15CWEE1061 pKa = 3.92AMIYY1065 pKa = 9.68WEE1067 pKa = 4.67VAAQTIYY1074 pKa = 10.86PEE1076 pKa = 4.55TVSHH1080 pKa = 6.63GSAVTTSARR1089 pKa = 11.84IPVSVMGQWFAPLYY1103 pKa = 8.59PQYY1106 pKa = 10.78FQLRR1110 pKa = 11.84LNLQVMSPQRR1120 pKa = 11.84TYY1122 pKa = 11.1ISINNVARR1130 pKa = 11.84AMFKK1134 pKa = 10.57VEE1136 pKa = 4.09YY1137 pKa = 10.61AEE1139 pKa = 4.18EE1140 pKa = 4.34TTGLLGKK1147 pKa = 10.05SLSPVSWLMWRR1158 pKa = 11.84LSLGVIISDD1167 pKa = 3.37TVSANNGWIVQCVNNGGVFDD1187 pKa = 4.68YY1188 pKa = 9.88YY1189 pKa = 10.66IHH1191 pKa = 6.66MEE1193 pKa = 4.05QPDD1196 pKa = 3.4ASRR1199 pKa = 11.84NQVMCRR1205 pKa = 11.84EE1206 pKa = 4.09YY1207 pKa = 10.77GAGNVTVPPTTKK1219 pKa = 9.87ISLPTGEE1226 pKa = 5.35FILKK1230 pKa = 10.3RR1231 pKa = 11.84SIATGLQPFSLLMQGPRR1248 pKa = 11.84DD1249 pKa = 3.55AFIGVGTGSTAYY1261 pKa = 10.88QMFTQTNTPQAIFVSEE1277 pKa = 4.26TWATIPDD1284 pKa = 4.01WLFSPAGLQSVRR1296 pKa = 11.84DD1297 pKa = 4.1SII1299 pKa = 4.59
Molecular weight: 140.15 kDa
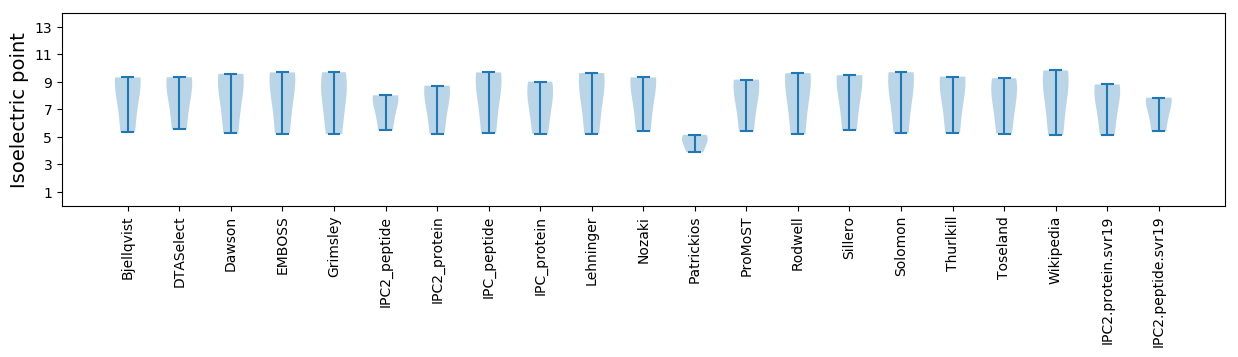
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF79|A0A1L3KF79_9VIRU RNA-directed RNA polymerase OS=Hubei toti-like virus 16 OX=1923304 PE=4 SV=1
MM1 pKa = 7.74TDD3 pKa = 3.84GLPGAVGDD11 pKa = 4.48SGSSGSGFRR20 pKa = 11.84RR21 pKa = 11.84FDD23 pKa = 3.22SFGIGSGQEE32 pKa = 3.72GDD34 pKa = 3.45RR35 pKa = 11.84GRR37 pKa = 11.84STGGSTDD44 pKa = 3.15RR45 pKa = 11.84TTYY48 pKa = 10.42FYY50 pKa = 11.1DD51 pKa = 3.42RR52 pKa = 11.84RR53 pKa = 11.84PDD55 pKa = 3.35GSVKK59 pKa = 9.54YY60 pKa = 9.2TEE62 pKa = 5.26LEE64 pKa = 3.78PATTIRR70 pKa = 11.84VDD72 pKa = 4.31GIKK75 pKa = 10.38DD76 pKa = 3.61SYY78 pKa = 11.8GSGSNSLLFAGSSRR92 pKa = 11.84TSLSSGVFEE101 pKa = 4.58STFGQLAFGSARR113 pKa = 11.84PLRR116 pKa = 11.84PLTDD120 pKa = 3.31AQRR123 pKa = 11.84RR124 pKa = 11.84SGVVTIEE131 pKa = 4.06RR132 pKa = 11.84NATPSTVHH140 pKa = 6.68RR141 pKa = 11.84PDD143 pKa = 3.14GHH145 pKa = 6.11HH146 pKa = 5.44QGSRR150 pKa = 11.84RR151 pKa = 11.84SRR153 pKa = 11.84LSRR156 pKa = 11.84FDD158 pKa = 4.0PLYY161 pKa = 10.75KK162 pKa = 9.04PTRR165 pKa = 11.84NKK167 pKa = 10.57RR168 pKa = 11.84SDD170 pKa = 3.4AGSAAARR177 pKa = 11.84DD178 pKa = 3.71EE179 pKa = 4.31SGILARR185 pKa = 11.84IAAGVASNFSKK196 pKa = 10.7SASPRR201 pKa = 11.84YY202 pKa = 9.1EE203 pKa = 3.71PRR205 pKa = 11.84SPTGSDD211 pKa = 3.01NEE213 pKa = 4.17FDD215 pKa = 5.01IIDD218 pKa = 4.11DD219 pKa = 4.2PRR221 pKa = 11.84DD222 pKa = 3.24EE223 pKa = 4.46SARR226 pKa = 11.84SASADD231 pKa = 3.46QQTSGG236 pKa = 3.77
MM1 pKa = 7.74TDD3 pKa = 3.84GLPGAVGDD11 pKa = 4.48SGSSGSGFRR20 pKa = 11.84RR21 pKa = 11.84FDD23 pKa = 3.22SFGIGSGQEE32 pKa = 3.72GDD34 pKa = 3.45RR35 pKa = 11.84GRR37 pKa = 11.84STGGSTDD44 pKa = 3.15RR45 pKa = 11.84TTYY48 pKa = 10.42FYY50 pKa = 11.1DD51 pKa = 3.42RR52 pKa = 11.84RR53 pKa = 11.84PDD55 pKa = 3.35GSVKK59 pKa = 9.54YY60 pKa = 9.2TEE62 pKa = 5.26LEE64 pKa = 3.78PATTIRR70 pKa = 11.84VDD72 pKa = 4.31GIKK75 pKa = 10.38DD76 pKa = 3.61SYY78 pKa = 11.8GSGSNSLLFAGSSRR92 pKa = 11.84TSLSSGVFEE101 pKa = 4.58STFGQLAFGSARR113 pKa = 11.84PLRR116 pKa = 11.84PLTDD120 pKa = 3.31AQRR123 pKa = 11.84RR124 pKa = 11.84SGVVTIEE131 pKa = 4.06RR132 pKa = 11.84NATPSTVHH140 pKa = 6.68RR141 pKa = 11.84PDD143 pKa = 3.14GHH145 pKa = 6.11HH146 pKa = 5.44QGSRR150 pKa = 11.84RR151 pKa = 11.84SRR153 pKa = 11.84LSRR156 pKa = 11.84FDD158 pKa = 4.0PLYY161 pKa = 10.75KK162 pKa = 9.04PTRR165 pKa = 11.84NKK167 pKa = 10.57RR168 pKa = 11.84SDD170 pKa = 3.4AGSAAARR177 pKa = 11.84DD178 pKa = 3.71EE179 pKa = 4.31SGILARR185 pKa = 11.84IAAGVASNFSKK196 pKa = 10.7SASPRR201 pKa = 11.84YY202 pKa = 9.1EE203 pKa = 3.71PRR205 pKa = 11.84SPTGSDD211 pKa = 3.01NEE213 pKa = 4.17FDD215 pKa = 5.01IIDD218 pKa = 4.11DD219 pKa = 4.2PRR221 pKa = 11.84DD222 pKa = 3.24EE223 pKa = 4.46SARR226 pKa = 11.84SASADD231 pKa = 3.46QQTSGG236 pKa = 3.77
Molecular weight: 25.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2394 |
236 |
1299 |
798.0 |
87.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.354 ± 1.418 | 1.42 ± 0.352 |
5.388 ± 1.088 | 4.094 ± 0.104 |
4.094 ± 0.093 | 8.062 ± 0.883 |
1.337 ± 0.083 | 5.597 ± 0.563 |
3.968 ± 0.817 | 8.855 ± 0.771 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.59 ± 0.614 | 4.135 ± 0.328 |
5.556 ± 0.923 | 4.637 ± 1.223 |
5.597 ± 1.646 | 9.148 ± 1.247 |
5.556 ± 0.924 | 7.185 ± 1.134 |
1.796 ± 0.29 | 2.632 ± 0.103 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
