
Hubei astro-like virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.65
Get precalculated fractions of proteins
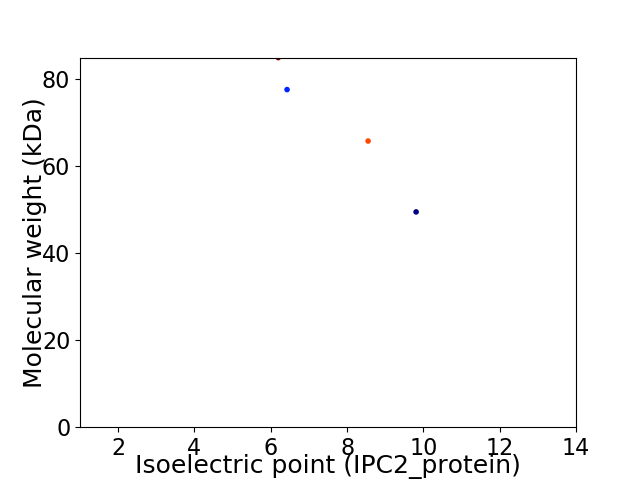
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KNZ3|A0A1L3KNZ3_9VIRU Capsid protein OS=Hubei astro-like virus OX=1922837 PE=4 SV=1
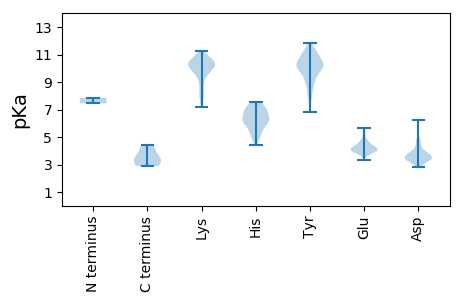
MM1 pKa = 7.49ANVGGTEE8 pKa = 3.76ARR10 pKa = 11.84RR11 pKa = 11.84EE12 pKa = 3.93PGGNGWVNIYY22 pKa = 10.65KK23 pKa = 10.43KK24 pKa = 10.53LKK26 pKa = 10.06KK27 pKa = 10.02ALKK30 pKa = 10.37DD31 pKa = 3.48PSSNQAVNLLSDD43 pKa = 4.35LVSGIEE49 pKa = 3.92QSLTAITRR57 pKa = 11.84EE58 pKa = 4.16TLMSTALSVLRR69 pKa = 11.84GKK71 pKa = 9.88LLNMVLTWIFFTIWVAFPYY90 pKa = 10.07VPRR93 pKa = 11.84VEE95 pKa = 4.79IDD97 pKa = 3.19LLLNYY102 pKa = 9.83DD103 pKa = 3.89LHH105 pKa = 7.41CFQLVEE111 pKa = 4.04NLAVVEE117 pKa = 4.08LVFRR121 pKa = 11.84AWFWYY126 pKa = 9.69KK127 pKa = 10.55LAFTFYY133 pKa = 10.1SLVYY137 pKa = 9.93FVTDD141 pKa = 3.51CLRR144 pKa = 11.84AYY146 pKa = 8.69WRR148 pKa = 11.84YY149 pKa = 8.9QSLRR153 pKa = 11.84EE154 pKa = 4.06LVWQVRR160 pKa = 11.84WTLMSCSNDD169 pKa = 2.9EE170 pKa = 5.6RR171 pKa = 11.84LDD173 pKa = 3.48DD174 pKa = 3.9WEE176 pKa = 5.07GMFDD180 pKa = 4.71LICSIFSPLCACAGTSAVNQWLTSYY205 pKa = 10.24TFGKK209 pKa = 10.43KK210 pKa = 9.91SFFATRR216 pKa = 11.84GASNFVQGYY225 pKa = 8.29IGRR228 pKa = 11.84NLGEE232 pKa = 4.17TVYY235 pKa = 10.77NLCVPRR241 pKa = 11.84AFQKK245 pKa = 10.54SDD247 pKa = 2.84CHH249 pKa = 6.21TWRR252 pKa = 11.84CWVRR256 pKa = 11.84PYY258 pKa = 10.64PGARR262 pKa = 11.84LLQHH266 pKa = 6.55CNHH269 pKa = 6.35RR270 pKa = 11.84MVLAEE275 pKa = 3.98QGVDD279 pKa = 2.98AADD282 pKa = 3.9FMSPGMFLLVPLMFVGWCVRR302 pKa = 11.84RR303 pKa = 11.84VFKK306 pKa = 10.16GTEE309 pKa = 3.83SRR311 pKa = 11.84QGLPSLTRR319 pKa = 11.84YY320 pKa = 10.22DD321 pKa = 3.83CLTPLRR327 pKa = 11.84GPTPEE332 pKa = 4.7SIFPGSPRR340 pKa = 11.84IGAVTKK346 pKa = 10.68AADD349 pKa = 3.03ATAAVLHH356 pKa = 6.68RR357 pKa = 11.84GTRR360 pKa = 11.84VATCFFVGNTIVTCKK375 pKa = 9.98HH376 pKa = 5.99AYY378 pKa = 9.75EE379 pKa = 4.51PDD381 pKa = 3.2EE382 pKa = 4.55EE383 pKa = 4.73GFHH386 pKa = 6.65FSFRR390 pKa = 11.84GVDD393 pKa = 3.32WTLKK397 pKa = 10.49SPVAKK402 pKa = 10.26FFPASVGCEE411 pKa = 3.94EE412 pKa = 4.23IVVVSMPKK420 pKa = 10.49DD421 pKa = 3.53LVSGLLNFGAMRR433 pKa = 11.84FKK435 pKa = 10.23TRR437 pKa = 11.84KK438 pKa = 9.73LVDD441 pKa = 3.24KK442 pKa = 9.79TAGGSIGYY450 pKa = 8.74SASNMYY456 pKa = 10.09HH457 pKa = 7.04LSFATGTDD465 pKa = 3.02IDD467 pKa = 4.37PDD469 pKa = 3.76GTHH472 pKa = 6.94NISSSPGSSGSPVFDD487 pKa = 4.29CNGIAVGVHH496 pKa = 5.11NAGGAVSNFFLPFSDD511 pKa = 4.55DD512 pKa = 3.65CIGYY516 pKa = 8.02MRR518 pKa = 11.84TTAPVDD524 pKa = 3.27GSVVVTAIHH533 pKa = 6.31GGWDD537 pKa = 3.2ATKK540 pKa = 8.72PTDD543 pKa = 3.03KK544 pKa = 10.17GTPVHH549 pKa = 6.61VLDD552 pKa = 5.22NVNTNLGSSSVGKK565 pKa = 9.86NDD567 pKa = 3.0AYY569 pKa = 9.69TRR571 pKa = 11.84CEE573 pKa = 4.0QFVSWATNSTNQQAWDD589 pKa = 3.67DD590 pKa = 3.8EE591 pKa = 4.8RR592 pKa = 11.84NPRR595 pKa = 11.84RR596 pKa = 11.84PKK598 pKa = 10.44AVPGTCASRR607 pKa = 11.84NAPVVSSVVTTVANNHH623 pKa = 6.05SGCNTVDD630 pKa = 3.85ADD632 pKa = 4.1HH633 pKa = 7.19SDD635 pKa = 3.15IFDD638 pKa = 3.5ARR640 pKa = 11.84VAQSIVSFYY649 pKa = 11.14KK650 pKa = 10.34EE651 pKa = 4.0YY652 pKa = 10.53TGPTLAAKK660 pKa = 10.24LDD662 pKa = 4.8AILHH666 pKa = 5.59SVLTDD671 pKa = 3.38HH672 pKa = 7.28FDD674 pKa = 3.46SDD676 pKa = 4.06MVDD679 pKa = 3.53KK680 pKa = 11.25LRR682 pKa = 11.84VAYY685 pKa = 9.39PGLNDD690 pKa = 3.47DD691 pKa = 4.91QIWARR696 pKa = 11.84ISKK699 pKa = 10.15RR700 pKa = 11.84CADD703 pKa = 3.75PGAIVVGSHH712 pKa = 5.27SQATAVVAALVEE724 pKa = 4.48GARR727 pKa = 11.84TIYY730 pKa = 9.96PDD732 pKa = 3.27EE733 pKa = 4.39WSDD736 pKa = 3.67EE737 pKa = 4.15YY738 pKa = 11.35LGSWLHH744 pKa = 5.91QRR746 pKa = 11.84YY747 pKa = 7.92PSCYY751 pKa = 7.75RR752 pKa = 11.84PANIPHH758 pKa = 6.11SVSDD762 pKa = 3.74QDD764 pKa = 4.4NQPACPNN771 pKa = 3.5
MM1 pKa = 7.49ANVGGTEE8 pKa = 3.76ARR10 pKa = 11.84RR11 pKa = 11.84EE12 pKa = 3.93PGGNGWVNIYY22 pKa = 10.65KK23 pKa = 10.43KK24 pKa = 10.53LKK26 pKa = 10.06KK27 pKa = 10.02ALKK30 pKa = 10.37DD31 pKa = 3.48PSSNQAVNLLSDD43 pKa = 4.35LVSGIEE49 pKa = 3.92QSLTAITRR57 pKa = 11.84EE58 pKa = 4.16TLMSTALSVLRR69 pKa = 11.84GKK71 pKa = 9.88LLNMVLTWIFFTIWVAFPYY90 pKa = 10.07VPRR93 pKa = 11.84VEE95 pKa = 4.79IDD97 pKa = 3.19LLLNYY102 pKa = 9.83DD103 pKa = 3.89LHH105 pKa = 7.41CFQLVEE111 pKa = 4.04NLAVVEE117 pKa = 4.08LVFRR121 pKa = 11.84AWFWYY126 pKa = 9.69KK127 pKa = 10.55LAFTFYY133 pKa = 10.1SLVYY137 pKa = 9.93FVTDD141 pKa = 3.51CLRR144 pKa = 11.84AYY146 pKa = 8.69WRR148 pKa = 11.84YY149 pKa = 8.9QSLRR153 pKa = 11.84EE154 pKa = 4.06LVWQVRR160 pKa = 11.84WTLMSCSNDD169 pKa = 2.9EE170 pKa = 5.6RR171 pKa = 11.84LDD173 pKa = 3.48DD174 pKa = 3.9WEE176 pKa = 5.07GMFDD180 pKa = 4.71LICSIFSPLCACAGTSAVNQWLTSYY205 pKa = 10.24TFGKK209 pKa = 10.43KK210 pKa = 9.91SFFATRR216 pKa = 11.84GASNFVQGYY225 pKa = 8.29IGRR228 pKa = 11.84NLGEE232 pKa = 4.17TVYY235 pKa = 10.77NLCVPRR241 pKa = 11.84AFQKK245 pKa = 10.54SDD247 pKa = 2.84CHH249 pKa = 6.21TWRR252 pKa = 11.84CWVRR256 pKa = 11.84PYY258 pKa = 10.64PGARR262 pKa = 11.84LLQHH266 pKa = 6.55CNHH269 pKa = 6.35RR270 pKa = 11.84MVLAEE275 pKa = 3.98QGVDD279 pKa = 2.98AADD282 pKa = 3.9FMSPGMFLLVPLMFVGWCVRR302 pKa = 11.84RR303 pKa = 11.84VFKK306 pKa = 10.16GTEE309 pKa = 3.83SRR311 pKa = 11.84QGLPSLTRR319 pKa = 11.84YY320 pKa = 10.22DD321 pKa = 3.83CLTPLRR327 pKa = 11.84GPTPEE332 pKa = 4.7SIFPGSPRR340 pKa = 11.84IGAVTKK346 pKa = 10.68AADD349 pKa = 3.03ATAAVLHH356 pKa = 6.68RR357 pKa = 11.84GTRR360 pKa = 11.84VATCFFVGNTIVTCKK375 pKa = 9.98HH376 pKa = 5.99AYY378 pKa = 9.75EE379 pKa = 4.51PDD381 pKa = 3.2EE382 pKa = 4.55EE383 pKa = 4.73GFHH386 pKa = 6.65FSFRR390 pKa = 11.84GVDD393 pKa = 3.32WTLKK397 pKa = 10.49SPVAKK402 pKa = 10.26FFPASVGCEE411 pKa = 3.94EE412 pKa = 4.23IVVVSMPKK420 pKa = 10.49DD421 pKa = 3.53LVSGLLNFGAMRR433 pKa = 11.84FKK435 pKa = 10.23TRR437 pKa = 11.84KK438 pKa = 9.73LVDD441 pKa = 3.24KK442 pKa = 9.79TAGGSIGYY450 pKa = 8.74SASNMYY456 pKa = 10.09HH457 pKa = 7.04LSFATGTDD465 pKa = 3.02IDD467 pKa = 4.37PDD469 pKa = 3.76GTHH472 pKa = 6.94NISSSPGSSGSPVFDD487 pKa = 4.29CNGIAVGVHH496 pKa = 5.11NAGGAVSNFFLPFSDD511 pKa = 4.55DD512 pKa = 3.65CIGYY516 pKa = 8.02MRR518 pKa = 11.84TTAPVDD524 pKa = 3.27GSVVVTAIHH533 pKa = 6.31GGWDD537 pKa = 3.2ATKK540 pKa = 8.72PTDD543 pKa = 3.03KK544 pKa = 10.17GTPVHH549 pKa = 6.61VLDD552 pKa = 5.22NVNTNLGSSSVGKK565 pKa = 9.86NDD567 pKa = 3.0AYY569 pKa = 9.69TRR571 pKa = 11.84CEE573 pKa = 4.0QFVSWATNSTNQQAWDD589 pKa = 3.67DD590 pKa = 3.8EE591 pKa = 4.8RR592 pKa = 11.84NPRR595 pKa = 11.84RR596 pKa = 11.84PKK598 pKa = 10.44AVPGTCASRR607 pKa = 11.84NAPVVSSVVTTVANNHH623 pKa = 6.05SGCNTVDD630 pKa = 3.85ADD632 pKa = 4.1HH633 pKa = 7.19SDD635 pKa = 3.15IFDD638 pKa = 3.5ARR640 pKa = 11.84VAQSIVSFYY649 pKa = 11.14KK650 pKa = 10.34EE651 pKa = 4.0YY652 pKa = 10.53TGPTLAAKK660 pKa = 10.24LDD662 pKa = 4.8AILHH666 pKa = 5.59SVLTDD671 pKa = 3.38HH672 pKa = 7.28FDD674 pKa = 3.46SDD676 pKa = 4.06MVDD679 pKa = 3.53KK680 pKa = 11.25LRR682 pKa = 11.84VAYY685 pKa = 9.39PGLNDD690 pKa = 3.47DD691 pKa = 4.91QIWARR696 pKa = 11.84ISKK699 pKa = 10.15RR700 pKa = 11.84CADD703 pKa = 3.75PGAIVVGSHH712 pKa = 5.27SQATAVVAALVEE724 pKa = 4.48GARR727 pKa = 11.84TIYY730 pKa = 9.96PDD732 pKa = 3.27EE733 pKa = 4.39WSDD736 pKa = 3.67EE737 pKa = 4.15YY738 pKa = 11.35LGSWLHH744 pKa = 5.91QRR746 pKa = 11.84YY747 pKa = 7.92PSCYY751 pKa = 7.75RR752 pKa = 11.84PANIPHH758 pKa = 6.11SVSDD762 pKa = 3.74QDD764 pKa = 4.4NQPACPNN771 pKa = 3.5
Molecular weight: 85.03 kDa
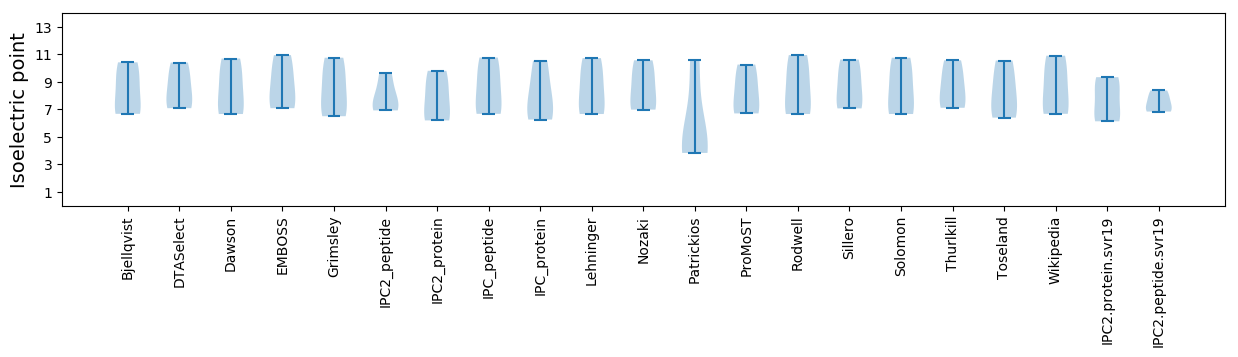
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KNT2|A0A1L3KNT2_9VIRU RNA-dependent RNA polymerase OS=Hubei astro-like virus OX=1922837 PE=4 SV=1
MM1 pKa = 7.8RR2 pKa = 11.84RR3 pKa = 11.84LLCYY7 pKa = 9.23TVALVLPLAFSLAILSSRR25 pKa = 11.84VSTRR29 pKa = 11.84MNRR32 pKa = 11.84TKK34 pKa = 10.38RR35 pKa = 11.84VSISLFAEE43 pKa = 4.71WIGLLSHH50 pKa = 7.02LWLSSFPLLLGVRR63 pKa = 11.84KK64 pKa = 9.92SLWFLCQKK72 pKa = 7.61TWCLVCSILVPCVLRR87 pKa = 11.84PGSWLIKK94 pKa = 9.87RR95 pKa = 11.84LVGQSVILLLTCTILVLLQEE115 pKa = 5.16PILTQMALITSLRR128 pKa = 11.84LPEE131 pKa = 4.12AVEE134 pKa = 4.1VLSSIVTGLPSVYY147 pKa = 10.32IMLAVRR153 pKa = 11.84SATSSYY159 pKa = 10.95PLVTTVLVTCVLLHH173 pKa = 6.21QLTDD177 pKa = 3.07QWLLPLFMVVGMPLSQLTKK196 pKa = 10.1VLQYY200 pKa = 9.39TYY202 pKa = 10.26WIMLIPIWGLHH213 pKa = 4.45LWGRR217 pKa = 11.84MMPTPDD223 pKa = 4.25ASSLYY228 pKa = 9.22PGPLIALTNKK238 pKa = 10.28LGTTSVIQEE247 pKa = 4.03GQRR250 pKa = 11.84LYY252 pKa = 11.11LEE254 pKa = 4.43PVQVEE259 pKa = 4.23MLLSYY264 pKa = 10.36PVLLLLWLITTLAVTLSMLTIRR286 pKa = 11.84TSLMRR291 pKa = 11.84VLHH294 pKa = 6.32RR295 pKa = 11.84ALSPFTKK302 pKa = 10.55NILVQPWLLSWTLFCTACSQITLIVTWSTSSAWLILGSMMTKK344 pKa = 10.55FGQGYY349 pKa = 9.31QSAALTLVRR358 pKa = 11.84LWWDD362 pKa = 3.57LTRR365 pKa = 11.84KK366 pKa = 8.62PRR368 pKa = 11.84LWLQRR373 pKa = 11.84WWKK376 pKa = 10.56GPGPFILMNGQTSTLGLGYY395 pKa = 9.31TKK397 pKa = 9.63GTQVAIVPRR406 pKa = 11.84TSHH409 pKa = 5.7TALAIKK415 pKa = 7.16TTNQPAQTRR424 pKa = 11.84PSRR427 pKa = 11.84HH428 pKa = 5.14GFRR431 pKa = 11.84PVKK434 pKa = 9.83FVSGGSTT441 pKa = 2.89
MM1 pKa = 7.8RR2 pKa = 11.84RR3 pKa = 11.84LLCYY7 pKa = 9.23TVALVLPLAFSLAILSSRR25 pKa = 11.84VSTRR29 pKa = 11.84MNRR32 pKa = 11.84TKK34 pKa = 10.38RR35 pKa = 11.84VSISLFAEE43 pKa = 4.71WIGLLSHH50 pKa = 7.02LWLSSFPLLLGVRR63 pKa = 11.84KK64 pKa = 9.92SLWFLCQKK72 pKa = 7.61TWCLVCSILVPCVLRR87 pKa = 11.84PGSWLIKK94 pKa = 9.87RR95 pKa = 11.84LVGQSVILLLTCTILVLLQEE115 pKa = 5.16PILTQMALITSLRR128 pKa = 11.84LPEE131 pKa = 4.12AVEE134 pKa = 4.1VLSSIVTGLPSVYY147 pKa = 10.32IMLAVRR153 pKa = 11.84SATSSYY159 pKa = 10.95PLVTTVLVTCVLLHH173 pKa = 6.21QLTDD177 pKa = 3.07QWLLPLFMVVGMPLSQLTKK196 pKa = 10.1VLQYY200 pKa = 9.39TYY202 pKa = 10.26WIMLIPIWGLHH213 pKa = 4.45LWGRR217 pKa = 11.84MMPTPDD223 pKa = 4.25ASSLYY228 pKa = 9.22PGPLIALTNKK238 pKa = 10.28LGTTSVIQEE247 pKa = 4.03GQRR250 pKa = 11.84LYY252 pKa = 11.11LEE254 pKa = 4.43PVQVEE259 pKa = 4.23MLLSYY264 pKa = 10.36PVLLLLWLITTLAVTLSMLTIRR286 pKa = 11.84TSLMRR291 pKa = 11.84VLHH294 pKa = 6.32RR295 pKa = 11.84ALSPFTKK302 pKa = 10.55NILVQPWLLSWTLFCTACSQITLIVTWSTSSAWLILGSMMTKK344 pKa = 10.55FGQGYY349 pKa = 9.31QSAALTLVRR358 pKa = 11.84LWWDD362 pKa = 3.57LTRR365 pKa = 11.84KK366 pKa = 8.62PRR368 pKa = 11.84LWLQRR373 pKa = 11.84WWKK376 pKa = 10.56GPGPFILMNGQTSTLGLGYY395 pKa = 9.31TKK397 pKa = 9.63GTQVAIVPRR406 pKa = 11.84TSHH409 pKa = 5.7TALAIKK415 pKa = 7.16TTNQPAQTRR424 pKa = 11.84PSRR427 pKa = 11.84HH428 pKa = 5.14GFRR431 pKa = 11.84PVKK434 pKa = 9.83FVSGGSTT441 pKa = 2.89
Molecular weight: 49.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2502 |
441 |
771 |
625.5 |
69.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.114 ± 0.778 | 1.918 ± 0.499 |
4.756 ± 0.823 | 3.597 ± 0.557 |
3.917 ± 0.397 | 6.875 ± 0.562 |
2.278 ± 0.362 | 4.836 ± 0.463 |
4.117 ± 0.711 | 9.592 ± 1.963 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.478 ± 0.272 | 3.597 ± 0.549 |
5.516 ± 0.381 | 3.637 ± 0.368 |
5.915 ± 0.567 | 7.874 ± 0.872 |
7.394 ± 0.96 | 7.634 ± 0.717 |
2.598 ± 0.483 | 3.357 ± 0.531 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
