
Coniophora puteana (strain RWD-64-598) (Brown rot fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Boletales; Coniophorineae; Coniophoraceae; Coniophora; Coniophora puteana
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
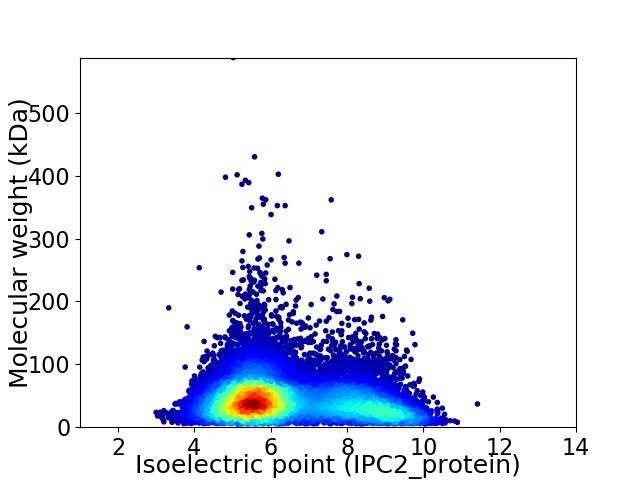
Virtual 2D-PAGE plot for 13735 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5M3MY85|A0A5M3MY85_CONPW Uncharacterized protein OS=Coniophora puteana (strain RWD-64-598) OX=741705 GN=CONPUDRAFT_150798 PE=4 SV=1
MM1 pKa = 8.02RR2 pKa = 11.84FTLATVIAVLPFLVDD17 pKa = 3.94AAPAPAARR25 pKa = 11.84APSNKK30 pKa = 9.19IAIQKK35 pKa = 10.26RR36 pKa = 11.84STLLNVDD43 pKa = 3.25GTVNLTALNRR53 pKa = 11.84HH54 pKa = 4.98TQSIKK59 pKa = 10.61AKK61 pKa = 8.43YY62 pKa = 9.14EE63 pKa = 3.96RR64 pKa = 11.84GFTNYY69 pKa = 10.15AKK71 pKa = 9.46NTGKK75 pKa = 9.58VHH77 pKa = 7.15SLAKK81 pKa = 10.36VNAGKK86 pKa = 10.29RR87 pKa = 11.84DD88 pKa = 3.51TGTVDD93 pKa = 3.27LTDD96 pKa = 4.61DD97 pKa = 5.1SSEE100 pKa = 3.67LWYY103 pKa = 10.88GKK105 pKa = 10.2ISVGTPAVEE114 pKa = 3.97YY115 pKa = 10.02TVDD118 pKa = 4.17FDD120 pKa = 4.24TGSSDD125 pKa = 4.51LFLPGPDD132 pKa = 4.81CDD134 pKa = 4.39DD135 pKa = 3.65TCSGHH140 pKa = 5.81TVYY143 pKa = 10.83DD144 pKa = 3.89PSKK147 pKa = 10.95SSDD150 pKa = 3.58SQDD153 pKa = 3.03VGQTFQLQYY162 pKa = 11.21GDD164 pKa = 3.63GSTVSGEE171 pKa = 3.82QWTDD175 pKa = 2.95TVTLAGLTATEE186 pKa = 4.13QTLGAAQQYY195 pKa = 7.87STGFEE200 pKa = 4.27SAQFPADD207 pKa = 3.13GLMGMAYY214 pKa = 9.41PAISSYY220 pKa = 10.72GASPVFNTLVSQGQTDD236 pKa = 3.24AGVFGFKK243 pKa = 9.26LTSSGAEE250 pKa = 3.83LTIGGVDD257 pKa = 3.47QSAVSGDD264 pKa = 3.46FTYY267 pKa = 11.07APVTQQGYY275 pKa = 5.89WQITADD281 pKa = 3.95GVSSGGQQGVGQFDD295 pKa = 5.29AIADD299 pKa = 3.8TGTTLIVGIPDD310 pKa = 3.69DD311 pKa = 3.57VASFYY316 pKa = 11.02EE317 pKa = 4.6AIGGQDD323 pKa = 3.14ASSTLGEE330 pKa = 4.27GYY332 pKa = 8.52YY333 pKa = 9.38TVPCDD338 pKa = 4.21SIPDD342 pKa = 3.55VTVTIGGQDD351 pKa = 3.62FPVSADD357 pKa = 3.48TFNLGAASQGSSDD370 pKa = 3.92CVGGIVGQDD379 pKa = 3.2LGQGFWILGDD389 pKa = 3.51VFLSNTYY396 pKa = 8.57TAFDD400 pKa = 3.84FDD402 pKa = 4.24NNQVGFAPLAA412 pKa = 4.1
MM1 pKa = 8.02RR2 pKa = 11.84FTLATVIAVLPFLVDD17 pKa = 3.94AAPAPAARR25 pKa = 11.84APSNKK30 pKa = 9.19IAIQKK35 pKa = 10.26RR36 pKa = 11.84STLLNVDD43 pKa = 3.25GTVNLTALNRR53 pKa = 11.84HH54 pKa = 4.98TQSIKK59 pKa = 10.61AKK61 pKa = 8.43YY62 pKa = 9.14EE63 pKa = 3.96RR64 pKa = 11.84GFTNYY69 pKa = 10.15AKK71 pKa = 9.46NTGKK75 pKa = 9.58VHH77 pKa = 7.15SLAKK81 pKa = 10.36VNAGKK86 pKa = 10.29RR87 pKa = 11.84DD88 pKa = 3.51TGTVDD93 pKa = 3.27LTDD96 pKa = 4.61DD97 pKa = 5.1SSEE100 pKa = 3.67LWYY103 pKa = 10.88GKK105 pKa = 10.2ISVGTPAVEE114 pKa = 3.97YY115 pKa = 10.02TVDD118 pKa = 4.17FDD120 pKa = 4.24TGSSDD125 pKa = 4.51LFLPGPDD132 pKa = 4.81CDD134 pKa = 4.39DD135 pKa = 3.65TCSGHH140 pKa = 5.81TVYY143 pKa = 10.83DD144 pKa = 3.89PSKK147 pKa = 10.95SSDD150 pKa = 3.58SQDD153 pKa = 3.03VGQTFQLQYY162 pKa = 11.21GDD164 pKa = 3.63GSTVSGEE171 pKa = 3.82QWTDD175 pKa = 2.95TVTLAGLTATEE186 pKa = 4.13QTLGAAQQYY195 pKa = 7.87STGFEE200 pKa = 4.27SAQFPADD207 pKa = 3.13GLMGMAYY214 pKa = 9.41PAISSYY220 pKa = 10.72GASPVFNTLVSQGQTDD236 pKa = 3.24AGVFGFKK243 pKa = 9.26LTSSGAEE250 pKa = 3.83LTIGGVDD257 pKa = 3.47QSAVSGDD264 pKa = 3.46FTYY267 pKa = 11.07APVTQQGYY275 pKa = 5.89WQITADD281 pKa = 3.95GVSSGGQQGVGQFDD295 pKa = 5.29AIADD299 pKa = 3.8TGTTLIVGIPDD310 pKa = 3.69DD311 pKa = 3.57VASFYY316 pKa = 11.02EE317 pKa = 4.6AIGGQDD323 pKa = 3.14ASSTLGEE330 pKa = 4.27GYY332 pKa = 8.52YY333 pKa = 9.38TVPCDD338 pKa = 4.21SIPDD342 pKa = 3.55VTVTIGGQDD351 pKa = 3.62FPVSADD357 pKa = 3.48TFNLGAASQGSSDD370 pKa = 3.92CVGGIVGQDD379 pKa = 3.2LGQGFWILGDD389 pKa = 3.51VFLSNTYY396 pKa = 8.57TAFDD400 pKa = 3.84FDD402 pKa = 4.24NNQVGFAPLAA412 pKa = 4.1
Molecular weight: 42.82 kDa
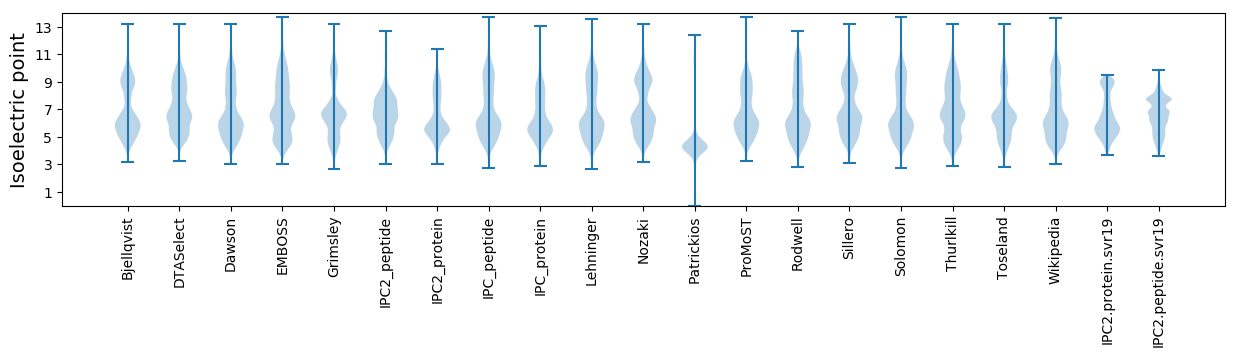
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5M3MII5|A0A5M3MII5_CONPW Uncharacterized protein OS=Coniophora puteana (strain RWD-64-598) OX=741705 GN=CONPUDRAFT_166778 PE=4 SV=1
MM1 pKa = 7.67IFNKK5 pKa = 10.43LSMLAVAAVMTAAVAAPVPEE25 pKa = 4.54PVTVRR30 pKa = 11.84APVTVRR36 pKa = 11.84SPVTVRR42 pKa = 11.84SPVTVRR48 pKa = 11.84APVTVRR54 pKa = 11.84SPVTVRR60 pKa = 11.84SPVTVRR66 pKa = 11.84SPVTVRR72 pKa = 11.84SPVTVRR78 pKa = 11.84SPVTVRR84 pKa = 11.84SPVTVRR90 pKa = 11.84SPVTVRR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84APVTVRR104 pKa = 11.84SPVTVRR110 pKa = 11.84SPVTVRR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84APVTVRR124 pKa = 11.84SPVTVRR130 pKa = 11.84SPVTVRR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84APVTVRR144 pKa = 11.84SPVTVRR150 pKa = 11.84SPVTVRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84APVTVRR164 pKa = 11.84SPVTVRR170 pKa = 11.84SPVTVRR176 pKa = 11.84SPVTVRR182 pKa = 11.84RR183 pKa = 11.84RR184 pKa = 11.84APVTVRR190 pKa = 11.84SPVTVRR196 pKa = 11.84APVTVRR202 pKa = 11.84SPVTVRR208 pKa = 11.84SPVTVRR214 pKa = 11.84SPVTVRR220 pKa = 11.84RR221 pKa = 11.84RR222 pKa = 11.84SPVTVRR228 pKa = 11.84SPVTVRR234 pKa = 11.84SPVTVRR240 pKa = 11.84RR241 pKa = 11.84RR242 pKa = 11.84APVTVRR248 pKa = 11.84SPVTVRR254 pKa = 11.84SPVTIRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84APVTVRR268 pKa = 11.84SPVTVRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84SPVTVRR282 pKa = 11.84SPVTVRR288 pKa = 11.84SPVTVRR294 pKa = 11.84SPVTVRR300 pKa = 11.84SPVTVRR306 pKa = 11.84SPVTVRR312 pKa = 11.84SPVTVRR318 pKa = 11.84SPVTVRR324 pKa = 11.84SPVTVRR330 pKa = 11.84APVTVRR336 pKa = 11.84AVEE339 pKa = 4.08LDD341 pKa = 3.25
MM1 pKa = 7.67IFNKK5 pKa = 10.43LSMLAVAAVMTAAVAAPVPEE25 pKa = 4.54PVTVRR30 pKa = 11.84APVTVRR36 pKa = 11.84SPVTVRR42 pKa = 11.84SPVTVRR48 pKa = 11.84APVTVRR54 pKa = 11.84SPVTVRR60 pKa = 11.84SPVTVRR66 pKa = 11.84SPVTVRR72 pKa = 11.84SPVTVRR78 pKa = 11.84SPVTVRR84 pKa = 11.84SPVTVRR90 pKa = 11.84SPVTVRR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84APVTVRR104 pKa = 11.84SPVTVRR110 pKa = 11.84SPVTVRR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84APVTVRR124 pKa = 11.84SPVTVRR130 pKa = 11.84SPVTVRR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84APVTVRR144 pKa = 11.84SPVTVRR150 pKa = 11.84SPVTVRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84APVTVRR164 pKa = 11.84SPVTVRR170 pKa = 11.84SPVTVRR176 pKa = 11.84SPVTVRR182 pKa = 11.84RR183 pKa = 11.84RR184 pKa = 11.84APVTVRR190 pKa = 11.84SPVTVRR196 pKa = 11.84APVTVRR202 pKa = 11.84SPVTVRR208 pKa = 11.84SPVTVRR214 pKa = 11.84SPVTVRR220 pKa = 11.84RR221 pKa = 11.84RR222 pKa = 11.84SPVTVRR228 pKa = 11.84SPVTVRR234 pKa = 11.84SPVTVRR240 pKa = 11.84RR241 pKa = 11.84RR242 pKa = 11.84APVTVRR248 pKa = 11.84SPVTVRR254 pKa = 11.84SPVTIRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84APVTVRR268 pKa = 11.84SPVTVRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84SPVTVRR282 pKa = 11.84SPVTVRR288 pKa = 11.84SPVTVRR294 pKa = 11.84SPVTVRR300 pKa = 11.84SPVTVRR306 pKa = 11.84SPVTVRR312 pKa = 11.84SPVTVRR318 pKa = 11.84SPVTVRR324 pKa = 11.84SPVTVRR330 pKa = 11.84APVTVRR336 pKa = 11.84AVEE339 pKa = 4.08LDD341 pKa = 3.25
Molecular weight: 36.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6006140 |
49 |
5304 |
437.3 |
48.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.064 ± 0.021 | 1.308 ± 0.009 |
5.703 ± 0.016 | 5.803 ± 0.021 |
3.646 ± 0.013 | 6.811 ± 0.023 |
2.604 ± 0.011 | 4.616 ± 0.018 |
4.194 ± 0.018 | 9.13 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.132 ± 0.008 | 3.346 ± 0.011 |
6.357 ± 0.025 | 3.692 ± 0.015 |
6.283 ± 0.021 | 8.667 ± 0.026 |
5.926 ± 0.013 | 6.506 ± 0.015 |
1.509 ± 0.008 | 2.705 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
