
Wenzhou crab virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Jingchuvirales; Chuviridae; Chuvivirus; Chuvivirus canceris
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
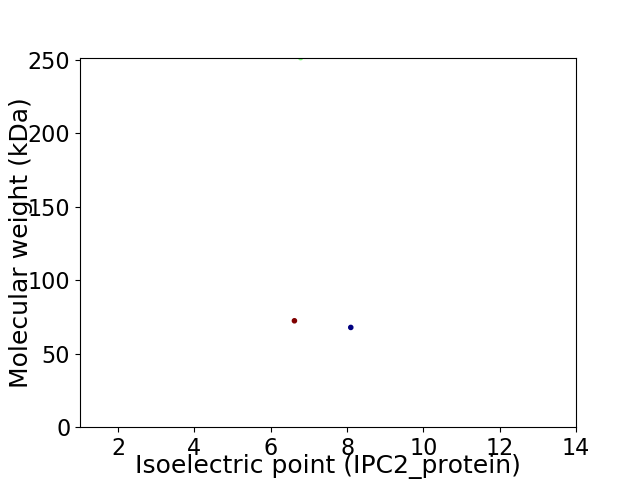
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KT97|A0A0B5KT97_9VIRU Putative nucleoprotein OS=Wenzhou crab virus 2 OX=1608092 GN=N PE=4 SV=1
MM1 pKa = 7.35VFFFLLVLCFARR13 pKa = 11.84GGLSAGPDD21 pKa = 3.62DD22 pKa = 4.09QVEE25 pKa = 3.99ALEE28 pKa = 5.59AYY30 pKa = 9.11DD31 pKa = 4.15CSGEE35 pKa = 4.1VSDD38 pKa = 5.72VSTISLNEE46 pKa = 3.77VKK48 pKa = 10.47EE49 pKa = 4.16CDD51 pKa = 3.52EE52 pKa = 4.34EE53 pKa = 4.27PTYY56 pKa = 11.1LEE58 pKa = 4.16SNQTIEE64 pKa = 4.17VQLLQDD70 pKa = 3.58SPNIVVDD77 pKa = 3.77VSTCRR82 pKa = 11.84LSISRR87 pKa = 11.84MIHH90 pKa = 5.09HH91 pKa = 7.5CGMHH95 pKa = 5.29SHH97 pKa = 6.7SSTVQAGMATYY108 pKa = 10.15VVEE111 pKa = 4.91LGRR114 pKa = 11.84SACLDD119 pKa = 3.23AVEE122 pKa = 4.35TRR124 pKa = 11.84RR125 pKa = 11.84VRR127 pKa = 11.84WSGTILDD134 pKa = 4.24GLSLGKK140 pKa = 8.39EE141 pKa = 4.04THH143 pKa = 6.04SLQTIAGSVTSGGKK157 pKa = 9.9CEE159 pKa = 3.93GSSYY163 pKa = 11.21SDD165 pKa = 3.9QFGSWSGVVVQAEE178 pKa = 3.74IKK180 pKa = 10.1MFIDD184 pKa = 4.87RR185 pKa = 11.84YY186 pKa = 9.42STTANIRR193 pKa = 11.84TGDD196 pKa = 3.17ITIRR200 pKa = 11.84GGLRR204 pKa = 11.84CEE206 pKa = 4.57WDD208 pKa = 3.51SQSCFDD214 pKa = 3.76NMAGEE219 pKa = 4.54TYY221 pKa = 8.95WRR223 pKa = 11.84KK224 pKa = 10.26KK225 pKa = 10.18EE226 pKa = 5.23DD227 pKa = 3.49GACPEE232 pKa = 4.0KK233 pKa = 11.12SFDD236 pKa = 4.01VLYY239 pKa = 10.34QGPAVRR245 pKa = 11.84VSSLRR250 pKa = 11.84LGTTHH255 pKa = 6.83HH256 pKa = 6.91MYY258 pKa = 10.35HH259 pKa = 7.06ISAGDD264 pKa = 3.59FQFVTQSGHH273 pKa = 5.52QDD275 pKa = 3.79YY276 pKa = 10.73LCHH279 pKa = 5.62QAVYY283 pKa = 8.66RR284 pKa = 11.84TDD286 pKa = 3.15HH287 pKa = 6.31PRR289 pKa = 11.84LYY291 pKa = 9.94IQEE294 pKa = 4.07RR295 pKa = 11.84PAPGFYY301 pKa = 10.12FKK303 pKa = 10.75KK304 pKa = 10.47KK305 pKa = 9.89SLQVRR310 pKa = 11.84NTDD313 pKa = 3.19LMQYY317 pKa = 9.69TNAKK321 pKa = 8.41LTLLEE326 pKa = 4.17HH327 pKa = 6.67HH328 pKa = 6.91FEE330 pKa = 4.21EE331 pKa = 5.34QLGQLYY337 pKa = 10.23QKK339 pKa = 10.76LRR341 pKa = 11.84LDD343 pKa = 3.22TCLAQRR349 pKa = 11.84TALQNKK355 pKa = 9.73LSLLKK360 pKa = 10.87LKK362 pKa = 10.42GSHH365 pKa = 6.47GGQHH369 pKa = 5.88SLGGRR374 pKa = 11.84GTKK377 pKa = 9.85TIVAGEE383 pKa = 4.09VAHH386 pKa = 6.73VIRR389 pKa = 11.84CRR391 pKa = 11.84RR392 pKa = 11.84VRR394 pKa = 11.84VTTRR398 pKa = 11.84PSSEE402 pKa = 4.63CYY404 pKa = 10.33AEE406 pKa = 4.96LPVTYY411 pKa = 9.98GEE413 pKa = 4.4KK414 pKa = 10.38AGCILPISRR423 pKa = 11.84IIVPHH428 pKa = 5.94CTPTPCSDD436 pKa = 5.39IIPQKK441 pKa = 9.8WKK443 pKa = 10.01IGRR446 pKa = 11.84RR447 pKa = 11.84WISLDD452 pKa = 3.04PKK454 pKa = 10.49PRR456 pKa = 11.84VTLPPVVLDD465 pKa = 3.87PAGKK469 pKa = 8.14TKK471 pKa = 10.18WEE473 pKa = 4.0YY474 pKa = 10.97KK475 pKa = 9.5PLRR478 pKa = 11.84EE479 pKa = 4.16LSIAGIYY486 pKa = 7.8STSQLEE492 pKa = 4.04QTQDD496 pKa = 3.33VIMNSHH502 pKa = 4.73TRR504 pKa = 11.84EE505 pKa = 4.07VVSSIISGRR514 pKa = 11.84VTGHH518 pKa = 7.25DD519 pKa = 3.92PDD521 pKa = 4.15DD522 pKa = 3.82QKK524 pKa = 11.83LAMSRR529 pKa = 11.84VIPMEE534 pKa = 3.66QLRR537 pKa = 11.84ASMSEE542 pKa = 3.8AFGTLTAWVLEE553 pKa = 4.36AGSAFGAVMLAYY565 pKa = 8.6YY566 pKa = 8.94TYY568 pKa = 10.65QAGKK572 pKa = 8.12TILSAVLNFKK582 pKa = 9.36MMRR585 pKa = 11.84EE586 pKa = 4.09MKK588 pKa = 10.44GCSAWLFTAPCSVVTLWLQHH608 pKa = 6.32RR609 pKa = 11.84SRR611 pKa = 11.84SSAGKK616 pKa = 9.65EE617 pKa = 3.89EE618 pKa = 4.43SCIGMVSEE626 pKa = 5.35AGPEE630 pKa = 4.06EE631 pKa = 4.3SRR633 pKa = 11.84LVHH636 pKa = 6.65PATIPGAVKK645 pKa = 10.05VYY647 pKa = 10.68PNVPEE652 pKa = 3.81QQ653 pKa = 3.2
MM1 pKa = 7.35VFFFLLVLCFARR13 pKa = 11.84GGLSAGPDD21 pKa = 3.62DD22 pKa = 4.09QVEE25 pKa = 3.99ALEE28 pKa = 5.59AYY30 pKa = 9.11DD31 pKa = 4.15CSGEE35 pKa = 4.1VSDD38 pKa = 5.72VSTISLNEE46 pKa = 3.77VKK48 pKa = 10.47EE49 pKa = 4.16CDD51 pKa = 3.52EE52 pKa = 4.34EE53 pKa = 4.27PTYY56 pKa = 11.1LEE58 pKa = 4.16SNQTIEE64 pKa = 4.17VQLLQDD70 pKa = 3.58SPNIVVDD77 pKa = 3.77VSTCRR82 pKa = 11.84LSISRR87 pKa = 11.84MIHH90 pKa = 5.09HH91 pKa = 7.5CGMHH95 pKa = 5.29SHH97 pKa = 6.7SSTVQAGMATYY108 pKa = 10.15VVEE111 pKa = 4.91LGRR114 pKa = 11.84SACLDD119 pKa = 3.23AVEE122 pKa = 4.35TRR124 pKa = 11.84RR125 pKa = 11.84VRR127 pKa = 11.84WSGTILDD134 pKa = 4.24GLSLGKK140 pKa = 8.39EE141 pKa = 4.04THH143 pKa = 6.04SLQTIAGSVTSGGKK157 pKa = 9.9CEE159 pKa = 3.93GSSYY163 pKa = 11.21SDD165 pKa = 3.9QFGSWSGVVVQAEE178 pKa = 3.74IKK180 pKa = 10.1MFIDD184 pKa = 4.87RR185 pKa = 11.84YY186 pKa = 9.42STTANIRR193 pKa = 11.84TGDD196 pKa = 3.17ITIRR200 pKa = 11.84GGLRR204 pKa = 11.84CEE206 pKa = 4.57WDD208 pKa = 3.51SQSCFDD214 pKa = 3.76NMAGEE219 pKa = 4.54TYY221 pKa = 8.95WRR223 pKa = 11.84KK224 pKa = 10.26KK225 pKa = 10.18EE226 pKa = 5.23DD227 pKa = 3.49GACPEE232 pKa = 4.0KK233 pKa = 11.12SFDD236 pKa = 4.01VLYY239 pKa = 10.34QGPAVRR245 pKa = 11.84VSSLRR250 pKa = 11.84LGTTHH255 pKa = 6.83HH256 pKa = 6.91MYY258 pKa = 10.35HH259 pKa = 7.06ISAGDD264 pKa = 3.59FQFVTQSGHH273 pKa = 5.52QDD275 pKa = 3.79YY276 pKa = 10.73LCHH279 pKa = 5.62QAVYY283 pKa = 8.66RR284 pKa = 11.84TDD286 pKa = 3.15HH287 pKa = 6.31PRR289 pKa = 11.84LYY291 pKa = 9.94IQEE294 pKa = 4.07RR295 pKa = 11.84PAPGFYY301 pKa = 10.12FKK303 pKa = 10.75KK304 pKa = 10.47KK305 pKa = 9.89SLQVRR310 pKa = 11.84NTDD313 pKa = 3.19LMQYY317 pKa = 9.69TNAKK321 pKa = 8.41LTLLEE326 pKa = 4.17HH327 pKa = 6.67HH328 pKa = 6.91FEE330 pKa = 4.21EE331 pKa = 5.34QLGQLYY337 pKa = 10.23QKK339 pKa = 10.76LRR341 pKa = 11.84LDD343 pKa = 3.22TCLAQRR349 pKa = 11.84TALQNKK355 pKa = 9.73LSLLKK360 pKa = 10.87LKK362 pKa = 10.42GSHH365 pKa = 6.47GGQHH369 pKa = 5.88SLGGRR374 pKa = 11.84GTKK377 pKa = 9.85TIVAGEE383 pKa = 4.09VAHH386 pKa = 6.73VIRR389 pKa = 11.84CRR391 pKa = 11.84RR392 pKa = 11.84VRR394 pKa = 11.84VTTRR398 pKa = 11.84PSSEE402 pKa = 4.63CYY404 pKa = 10.33AEE406 pKa = 4.96LPVTYY411 pKa = 9.98GEE413 pKa = 4.4KK414 pKa = 10.38AGCILPISRR423 pKa = 11.84IIVPHH428 pKa = 5.94CTPTPCSDD436 pKa = 5.39IIPQKK441 pKa = 9.8WKK443 pKa = 10.01IGRR446 pKa = 11.84RR447 pKa = 11.84WISLDD452 pKa = 3.04PKK454 pKa = 10.49PRR456 pKa = 11.84VTLPPVVLDD465 pKa = 3.87PAGKK469 pKa = 8.14TKK471 pKa = 10.18WEE473 pKa = 4.0YY474 pKa = 10.97KK475 pKa = 9.5PLRR478 pKa = 11.84EE479 pKa = 4.16LSIAGIYY486 pKa = 7.8STSQLEE492 pKa = 4.04QTQDD496 pKa = 3.33VIMNSHH502 pKa = 4.73TRR504 pKa = 11.84EE505 pKa = 4.07VVSSIISGRR514 pKa = 11.84VTGHH518 pKa = 7.25DD519 pKa = 3.92PDD521 pKa = 4.15DD522 pKa = 3.82QKK524 pKa = 11.83LAMSRR529 pKa = 11.84VIPMEE534 pKa = 3.66QLRR537 pKa = 11.84ASMSEE542 pKa = 3.8AFGTLTAWVLEE553 pKa = 4.36AGSAFGAVMLAYY565 pKa = 8.6YY566 pKa = 8.94TYY568 pKa = 10.65QAGKK572 pKa = 8.12TILSAVLNFKK582 pKa = 9.36MMRR585 pKa = 11.84EE586 pKa = 4.09MKK588 pKa = 10.44GCSAWLFTAPCSVVTLWLQHH608 pKa = 6.32RR609 pKa = 11.84SRR611 pKa = 11.84SSAGKK616 pKa = 9.65EE617 pKa = 3.89EE618 pKa = 4.43SCIGMVSEE626 pKa = 5.35AGPEE630 pKa = 4.06EE631 pKa = 4.3SRR633 pKa = 11.84LVHH636 pKa = 6.65PATIPGAVKK645 pKa = 10.05VYY647 pKa = 10.68PNVPEE652 pKa = 3.81QQ653 pKa = 3.2
Molecular weight: 72.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KT97|A0A0B5KT97_9VIRU Putative nucleoprotein OS=Wenzhou crab virus 2 OX=1608092 GN=N PE=4 SV=1
MM1 pKa = 7.53EE2 pKa = 5.67KK3 pKa = 10.24LVPYY7 pKa = 7.88EE8 pKa = 3.8FPIRR12 pKa = 11.84RR13 pKa = 11.84GPHH16 pKa = 5.8PPYY19 pKa = 10.89GPMEE23 pKa = 4.35IEE25 pKa = 4.59NEE27 pKa = 4.07DD28 pKa = 3.63STTINPPAGLKK39 pKa = 10.3EE40 pKa = 4.25SIEE43 pKa = 4.25VFFLSYY49 pKa = 11.07QEE51 pKa = 4.43VLLTTTIPTTRR62 pKa = 11.84SGAKK66 pKa = 9.65KK67 pKa = 8.57GTRR70 pKa = 11.84KK71 pKa = 10.4GKK73 pKa = 8.58VTMSQQASSQTAGPQAPAAPSPAPVAQGTGNGTTPGNVIQGGSAPPALNAPGPQSLQAPPQTSASVHH140 pKa = 6.41PPTPVPSTAGQAPILAPASSAPYY163 pKa = 9.67QGPTFIPAGSSQLVQMTHH181 pKa = 6.44PVLPVHH187 pKa = 6.26IQGAGFNPMYY197 pKa = 10.28QGFYY201 pKa = 7.7NTPMATAAGPSQVAYY216 pKa = 9.46PGAGWPSQPQNNPGAQAPPAPAFTFNAGPASNKK249 pKa = 7.39VQWGVAFGRR258 pKa = 11.84DD259 pKa = 3.29LKK261 pKa = 11.34YY262 pKa = 11.04DD263 pKa = 3.73AFRR266 pKa = 11.84LKK268 pKa = 10.64NLNHH272 pKa = 6.6GVFQGAWDD280 pKa = 4.15DD281 pKa = 4.45ADD283 pKa = 3.9LLLYY287 pKa = 9.89VAFYY291 pKa = 11.03GRR293 pKa = 11.84VRR295 pKa = 11.84AQVVGTNMMYY305 pKa = 10.83AVTIFAIGEE314 pKa = 4.4GIMDD318 pKa = 5.35CLPDD322 pKa = 4.5SAYY325 pKa = 10.96LDD327 pKa = 3.57QAQAQAAFIQDD338 pKa = 3.21ISSLFPLSSVGWLRR352 pKa = 11.84VDD354 pKa = 2.84ATTYY358 pKa = 11.61ANALTNEE365 pKa = 3.93LRR367 pKa = 11.84GLNLPATHH375 pKa = 6.83SEE377 pKa = 4.13GLAQALDD384 pKa = 3.58LRR386 pKa = 11.84GAYY389 pKa = 10.04AVMVWLAKK397 pKa = 10.42RR398 pKa = 11.84LTASVRR404 pKa = 11.84KK405 pKa = 8.23TEE407 pKa = 4.45IILWTLVALVKK418 pKa = 10.5QGNISDD424 pKa = 3.93RR425 pKa = 11.84FTNKK429 pKa = 8.02ITQSLEE435 pKa = 3.74QEE437 pKa = 4.26VGVTVTLPSDD447 pKa = 4.65GIRR450 pKa = 11.84AVWDD454 pKa = 3.61VYY456 pKa = 11.13GPYY459 pKa = 9.94INEE462 pKa = 3.86VTAGPVFNRR471 pKa = 11.84WLPLVPQNALRR482 pKa = 11.84VRR484 pKa = 11.84LTILQAAGEE493 pKa = 4.33GLTAFTTILKK503 pKa = 10.22ALRR506 pKa = 11.84AHH508 pKa = 7.0PRR510 pKa = 11.84FHH512 pKa = 7.01WSRR515 pKa = 11.84VINLFPQEE523 pKa = 3.27WDD525 pKa = 3.27NFVNAVTAVGANTFYY540 pKa = 11.19GFKK543 pKa = 10.15KK544 pKa = 10.36DD545 pKa = 3.56LGVVRR550 pKa = 11.84STMYY554 pKa = 11.09KK555 pKa = 9.64NLGYY559 pKa = 9.76VAKK562 pKa = 9.8EE563 pKa = 3.7LMVKK567 pKa = 10.56VSGDD571 pKa = 3.35APLNFYY577 pKa = 10.88AGWTRR582 pKa = 11.84IPAFRR587 pKa = 11.84TIIDD591 pKa = 3.6QMVVDD596 pKa = 4.94YY597 pKa = 9.25EE598 pKa = 4.21AQRR601 pKa = 11.84AADD604 pKa = 3.79LTGGGHH610 pKa = 7.26AAYY613 pKa = 10.39LPTADD618 pKa = 4.77AALSTPAIMSSTFPLNN634 pKa = 3.07
MM1 pKa = 7.53EE2 pKa = 5.67KK3 pKa = 10.24LVPYY7 pKa = 7.88EE8 pKa = 3.8FPIRR12 pKa = 11.84RR13 pKa = 11.84GPHH16 pKa = 5.8PPYY19 pKa = 10.89GPMEE23 pKa = 4.35IEE25 pKa = 4.59NEE27 pKa = 4.07DD28 pKa = 3.63STTINPPAGLKK39 pKa = 10.3EE40 pKa = 4.25SIEE43 pKa = 4.25VFFLSYY49 pKa = 11.07QEE51 pKa = 4.43VLLTTTIPTTRR62 pKa = 11.84SGAKK66 pKa = 9.65KK67 pKa = 8.57GTRR70 pKa = 11.84KK71 pKa = 10.4GKK73 pKa = 8.58VTMSQQASSQTAGPQAPAAPSPAPVAQGTGNGTTPGNVIQGGSAPPALNAPGPQSLQAPPQTSASVHH140 pKa = 6.41PPTPVPSTAGQAPILAPASSAPYY163 pKa = 9.67QGPTFIPAGSSQLVQMTHH181 pKa = 6.44PVLPVHH187 pKa = 6.26IQGAGFNPMYY197 pKa = 10.28QGFYY201 pKa = 7.7NTPMATAAGPSQVAYY216 pKa = 9.46PGAGWPSQPQNNPGAQAPPAPAFTFNAGPASNKK249 pKa = 7.39VQWGVAFGRR258 pKa = 11.84DD259 pKa = 3.29LKK261 pKa = 11.34YY262 pKa = 11.04DD263 pKa = 3.73AFRR266 pKa = 11.84LKK268 pKa = 10.64NLNHH272 pKa = 6.6GVFQGAWDD280 pKa = 4.15DD281 pKa = 4.45ADD283 pKa = 3.9LLLYY287 pKa = 9.89VAFYY291 pKa = 11.03GRR293 pKa = 11.84VRR295 pKa = 11.84AQVVGTNMMYY305 pKa = 10.83AVTIFAIGEE314 pKa = 4.4GIMDD318 pKa = 5.35CLPDD322 pKa = 4.5SAYY325 pKa = 10.96LDD327 pKa = 3.57QAQAQAAFIQDD338 pKa = 3.21ISSLFPLSSVGWLRR352 pKa = 11.84VDD354 pKa = 2.84ATTYY358 pKa = 11.61ANALTNEE365 pKa = 3.93LRR367 pKa = 11.84GLNLPATHH375 pKa = 6.83SEE377 pKa = 4.13GLAQALDD384 pKa = 3.58LRR386 pKa = 11.84GAYY389 pKa = 10.04AVMVWLAKK397 pKa = 10.42RR398 pKa = 11.84LTASVRR404 pKa = 11.84KK405 pKa = 8.23TEE407 pKa = 4.45IILWTLVALVKK418 pKa = 10.5QGNISDD424 pKa = 3.93RR425 pKa = 11.84FTNKK429 pKa = 8.02ITQSLEE435 pKa = 3.74QEE437 pKa = 4.26VGVTVTLPSDD447 pKa = 4.65GIRR450 pKa = 11.84AVWDD454 pKa = 3.61VYY456 pKa = 11.13GPYY459 pKa = 9.94INEE462 pKa = 3.86VTAGPVFNRR471 pKa = 11.84WLPLVPQNALRR482 pKa = 11.84VRR484 pKa = 11.84LTILQAAGEE493 pKa = 4.33GLTAFTTILKK503 pKa = 10.22ALRR506 pKa = 11.84AHH508 pKa = 7.0PRR510 pKa = 11.84FHH512 pKa = 7.01WSRR515 pKa = 11.84VINLFPQEE523 pKa = 3.27WDD525 pKa = 3.27NFVNAVTAVGANTFYY540 pKa = 11.19GFKK543 pKa = 10.15KK544 pKa = 10.36DD545 pKa = 3.56LGVVRR550 pKa = 11.84STMYY554 pKa = 11.09KK555 pKa = 9.64NLGYY559 pKa = 9.76VAKK562 pKa = 9.8EE563 pKa = 3.7LMVKK567 pKa = 10.56VSGDD571 pKa = 3.35APLNFYY577 pKa = 10.88AGWTRR582 pKa = 11.84IPAFRR587 pKa = 11.84TIIDD591 pKa = 3.6QMVVDD596 pKa = 4.94YY597 pKa = 9.25EE598 pKa = 4.21AQRR601 pKa = 11.84AADD604 pKa = 3.79LTGGGHH610 pKa = 7.26AAYY613 pKa = 10.39LPTADD618 pKa = 4.77AALSTPAIMSSTFPLNN634 pKa = 3.07
Molecular weight: 67.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3495 |
634 |
2208 |
1165.0 |
130.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.754 ± 1.46 | 1.888 ± 0.541 |
4.578 ± 0.421 | 5.522 ± 0.763 |
3.72 ± 0.364 | 6.094 ± 1.114 |
2.918 ± 0.458 | 4.835 ± 0.171 |
4.578 ± 0.545 | 9.986 ± 0.884 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.804 ± 0.237 | 3.348 ± 0.556 |
5.608 ± 1.053 | 4.435 ± 0.506 |
5.608 ± 0.505 | 7.382 ± 0.682 |
6.552 ± 0.404 | 6.981 ± 0.331 |
1.574 ± 0.049 | 3.834 ± 0.245 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
