
Sediminicola luteus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Sediminicola
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
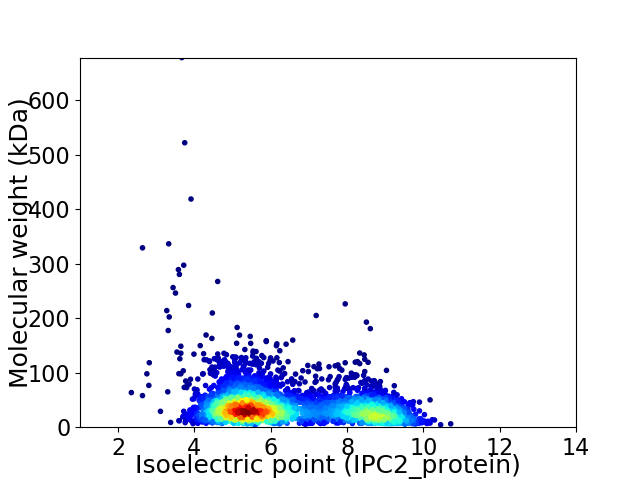
Virtual 2D-PAGE plot for 3771 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A4GAC9|A0A2A4GAC9_9FLAO Galactokinase OS=Sediminicola luteus OX=319238 GN=B7P33_05890 PE=3 SV=1
MM1 pKa = 7.4EE2 pKa = 5.18SKK4 pKa = 10.39EE5 pKa = 3.94KK6 pKa = 10.82AMKK9 pKa = 9.97KK10 pKa = 10.4LNISTYY16 pKa = 10.49VIALVLLVLALAVFMGCEE34 pKa = 4.36PDD36 pKa = 5.11DD37 pKa = 4.4SDD39 pKa = 5.47AFPASLPDD47 pKa = 3.45TAAVFTDD54 pKa = 4.6GFSPGLDD61 pKa = 3.13YY62 pKa = 10.89TAFGDD67 pKa = 4.01SKK69 pKa = 10.4VTAFQLDD76 pKa = 3.76EE77 pKa = 4.13EE78 pKa = 4.62VTYY81 pKa = 10.92DD82 pKa = 3.88KK83 pKa = 11.1SDD85 pKa = 2.83SAMRR89 pKa = 11.84FDD91 pKa = 5.49IPNQGDD97 pKa = 3.89PEE99 pKa = 4.24SGYY102 pKa = 11.37AGGIFFDD109 pKa = 3.31KK110 pKa = 10.5TGRR113 pKa = 11.84NLSGYY118 pKa = 9.89NVLSFWGKK126 pKa = 9.88ASQGAIIQEE135 pKa = 4.31LGFGLTDD142 pKa = 3.2TQMYY146 pKa = 6.86RR147 pKa = 11.84TSITDD152 pKa = 3.38VAFSTAWTKK161 pKa = 11.06YY162 pKa = 10.25YY163 pKa = 10.33ILLPDD168 pKa = 4.18PARR171 pKa = 11.84LTMEE175 pKa = 3.78EE176 pKa = 4.28GMFYY180 pKa = 10.59FVDD183 pKa = 3.81TPDD186 pKa = 3.24NGKK189 pKa = 10.73GYY191 pKa = 9.99SFWIDD196 pKa = 3.06EE197 pKa = 4.32LRR199 pKa = 11.84FEE201 pKa = 4.34NSGTIGQVQAYY212 pKa = 7.73IQNGEE217 pKa = 4.35EE218 pKa = 4.07VTSTVFVGQEE228 pKa = 3.99VPVSGLGVSFSLPNGVSQAVNAAPAFFTLASSNPQVAQVVVDD270 pKa = 3.61EE271 pKa = 4.55TTQVSTVRR279 pKa = 11.84ILAQGEE285 pKa = 4.2AVITASMGDD294 pKa = 3.66VAAMGSLTLQVRR306 pKa = 11.84EE307 pKa = 4.59FEE309 pKa = 4.41PAPLPDD315 pKa = 3.99KK316 pKa = 11.06VADD319 pKa = 3.87DD320 pKa = 4.54VISIFSDD327 pKa = 3.34AYY329 pKa = 9.32TNVPVDD335 pKa = 3.97FYY337 pKa = 11.44NGYY340 pKa = 9.09YY341 pKa = 10.63APFQTTVSADD351 pKa = 3.39FTVDD355 pKa = 3.05GDD357 pKa = 4.24HH358 pKa = 6.34VLNYY362 pKa = 10.05INYY365 pKa = 8.29NFVGIEE371 pKa = 3.99FNKK374 pKa = 10.07EE375 pKa = 3.72VPTIDD380 pKa = 3.42ATEE383 pKa = 4.08MTHH386 pKa = 6.19LHH388 pKa = 5.27VDD390 pKa = 3.55IYY392 pKa = 10.7IPEE395 pKa = 4.4ALDD398 pKa = 4.25PNATWRR404 pKa = 11.84VNLRR408 pKa = 11.84DD409 pKa = 4.25FGADD413 pKa = 3.27GNFDD417 pKa = 3.99GGDD420 pKa = 3.53DD421 pKa = 4.27TIASQLLTTASDD433 pKa = 3.64PALVSEE439 pKa = 4.26QWISVDD445 pKa = 2.97MDD447 pKa = 3.04IRR449 pKa = 11.84DD450 pKa = 3.91MANKK454 pKa = 8.38ATLGQIVFDD463 pKa = 4.57AEE465 pKa = 4.79GDD467 pKa = 3.72TSPRR471 pKa = 11.84PSGFYY476 pKa = 10.51VDD478 pKa = 5.44NLYY481 pKa = 10.86LYY483 pKa = 10.38NDD485 pKa = 3.48QGTGGGGPQPIPPTTAAPTPTLDD508 pKa = 3.17AAQVVSIFSDD518 pKa = 3.58AYY520 pKa = 9.95TDD522 pKa = 3.82VPNQGFNQYY531 pKa = 9.31GAATFAEE538 pKa = 4.66IPVADD543 pKa = 4.23NATLNYY549 pKa = 9.04KK550 pKa = 10.35QSDD553 pKa = 3.83NPGGNFQVIEE563 pKa = 4.47LGAEE567 pKa = 3.95NQIDD571 pKa = 3.9ATAMEE576 pKa = 4.64LTDD579 pKa = 3.72FHH581 pKa = 8.03VDD583 pKa = 2.64LWFPNPVNEE592 pKa = 4.07NTAFLLKK599 pKa = 10.24VVNIAPDD606 pKa = 3.33ITSEE610 pKa = 3.79ALIRR614 pKa = 11.84IDD616 pKa = 5.23ANATPAMAQGQWLAIDD632 pKa = 3.94LTMDD636 pKa = 3.3QLQAAGLAANSNIQQVVIDD655 pKa = 4.87LLDD658 pKa = 4.13AGEE661 pKa = 4.27VYY663 pKa = 10.34IDD665 pKa = 3.4NLYY668 pKa = 10.45FSKK671 pKa = 11.26SNGAGGSAPTTSAPEE686 pKa = 3.92PPARR690 pKa = 11.84DD691 pKa = 3.32AADD694 pKa = 3.66VISIFGEE701 pKa = 4.22AYY703 pKa = 9.89TNITGIDD710 pKa = 3.83YY711 pKa = 10.72DD712 pKa = 4.56PNWGQSGHH720 pKa = 5.26MQVNTAFDD728 pKa = 3.68SGDD731 pKa = 3.45GNLALAYY738 pKa = 9.71PSFNYY743 pKa = 10.11QGTDD747 pKa = 3.54FSGNAQNASEE757 pKa = 4.28MEE759 pKa = 4.21FLHH762 pKa = 7.81VDD764 pKa = 3.06IWVPAGTDD772 pKa = 3.22RR773 pKa = 11.84QVKK776 pKa = 9.6VSPINNGTGEE786 pKa = 4.33GEE788 pKa = 4.4VLVSVPLIPGSWNSVDD804 pKa = 5.51LPIADD809 pKa = 3.98FTGMTWDD816 pKa = 3.94SVVQLKK822 pKa = 10.5FDD824 pKa = 3.7GQFNGDD830 pKa = 3.11GSANTDD836 pKa = 3.64PFDD839 pKa = 3.62MYY841 pKa = 11.25LDD843 pKa = 3.25NVYY846 pKa = 10.17FYY848 pKa = 10.44KK849 pKa = 10.75EE850 pKa = 3.86PSAPSAAPTTSAPEE864 pKa = 3.83PLARR868 pKa = 11.84DD869 pKa = 3.38AADD872 pKa = 3.59VISIFGEE879 pKa = 4.22AYY881 pKa = 9.89TNITGIDD888 pKa = 3.83YY889 pKa = 10.72DD890 pKa = 4.56PNWGQSGHH898 pKa = 5.26MQVNTAFDD906 pKa = 4.3PGDD909 pKa = 3.75GNLALAYY916 pKa = 9.71PSFNYY921 pKa = 10.11QGTDD925 pKa = 3.54FSGNAQNASEE935 pKa = 4.28MEE937 pKa = 4.21FLHH940 pKa = 7.81VDD942 pKa = 3.06IWVPAGTDD950 pKa = 3.22RR951 pKa = 11.84QVKK954 pKa = 9.6VSPINNGTGEE964 pKa = 4.33GEE966 pKa = 4.4VLVSVPLTPGAWNSVDD982 pKa = 5.21LPMADD987 pKa = 3.63FTGMTWDD994 pKa = 3.94SVVQLKK1000 pKa = 10.5FDD1002 pKa = 3.7GQFNGDD1008 pKa = 3.11GSANTDD1014 pKa = 3.31PFDD1017 pKa = 3.99IYY1019 pKa = 11.05LDD1021 pKa = 3.23NVYY1024 pKa = 10.14FYY1026 pKa = 10.44KK1027 pKa = 10.7EE1028 pKa = 3.75PSAPSTAPTTSAPAPPVRR1046 pKa = 11.84DD1047 pKa = 3.33AADD1050 pKa = 3.61VISIFGEE1057 pKa = 4.22AYY1059 pKa = 9.89TNITGIDD1066 pKa = 3.83YY1067 pKa = 10.72DD1068 pKa = 4.56PNWGQSGHH1076 pKa = 5.26MQVNTAFDD1084 pKa = 4.3PGDD1087 pKa = 3.75GNLALAYY1094 pKa = 9.71PSFNYY1099 pKa = 10.11QGTDD1103 pKa = 3.54FSGNAQNAATMQFLHH1118 pKa = 7.2VDD1120 pKa = 3.2IWVPAGTDD1128 pKa = 3.22RR1129 pKa = 11.84QVKK1132 pKa = 9.6VSPINNGTGAGEE1144 pKa = 4.21VLVSVPLTPGAWNSVDD1160 pKa = 4.95LPIGDD1165 pKa = 4.06FTGMTWDD1172 pKa = 4.28SVFQLKK1178 pKa = 10.32FDD1180 pKa = 3.85GQFNGDD1186 pKa = 3.11GSANTDD1192 pKa = 3.31PFDD1195 pKa = 3.99IYY1197 pKa = 11.05LDD1199 pKa = 3.23NVYY1202 pKa = 10.14FYY1204 pKa = 10.44KK1205 pKa = 10.7EE1206 pKa = 3.75PSAPSTAPTTSAPAPPVRR1224 pKa = 11.84DD1225 pKa = 3.33AADD1228 pKa = 3.61VISIFGEE1235 pKa = 4.22AYY1237 pKa = 9.67TNITGVDD1244 pKa = 3.77YY1245 pKa = 11.05DD1246 pKa = 4.63PNWGQSGHH1254 pKa = 5.26MQVNTAFDD1262 pKa = 4.3PGDD1265 pKa = 3.75GNLALAYY1272 pKa = 9.89PNFNYY1277 pKa = 10.28QGTDD1281 pKa = 3.53FSGNAQNAASMQFLHH1296 pKa = 7.39VDD1298 pKa = 3.07IWVPAGTDD1306 pKa = 3.22RR1307 pKa = 11.84QVKK1310 pKa = 9.6VSPINNGTGAGEE1322 pKa = 4.21VLVSVPLTPGAWNSVDD1338 pKa = 4.95LPIGDD1343 pKa = 4.06FTGMTWDD1350 pKa = 4.28SVFQLKK1356 pKa = 10.32FDD1358 pKa = 3.85GQFNGDD1364 pKa = 3.11GSANTDD1370 pKa = 3.42PFDD1373 pKa = 3.73VYY1375 pKa = 11.29LDD1377 pKa = 3.31NVYY1380 pKa = 10.22FYY1382 pKa = 11.57KK1383 pKa = 10.9NN1384 pKa = 3.04
MM1 pKa = 7.4EE2 pKa = 5.18SKK4 pKa = 10.39EE5 pKa = 3.94KK6 pKa = 10.82AMKK9 pKa = 9.97KK10 pKa = 10.4LNISTYY16 pKa = 10.49VIALVLLVLALAVFMGCEE34 pKa = 4.36PDD36 pKa = 5.11DD37 pKa = 4.4SDD39 pKa = 5.47AFPASLPDD47 pKa = 3.45TAAVFTDD54 pKa = 4.6GFSPGLDD61 pKa = 3.13YY62 pKa = 10.89TAFGDD67 pKa = 4.01SKK69 pKa = 10.4VTAFQLDD76 pKa = 3.76EE77 pKa = 4.13EE78 pKa = 4.62VTYY81 pKa = 10.92DD82 pKa = 3.88KK83 pKa = 11.1SDD85 pKa = 2.83SAMRR89 pKa = 11.84FDD91 pKa = 5.49IPNQGDD97 pKa = 3.89PEE99 pKa = 4.24SGYY102 pKa = 11.37AGGIFFDD109 pKa = 3.31KK110 pKa = 10.5TGRR113 pKa = 11.84NLSGYY118 pKa = 9.89NVLSFWGKK126 pKa = 9.88ASQGAIIQEE135 pKa = 4.31LGFGLTDD142 pKa = 3.2TQMYY146 pKa = 6.86RR147 pKa = 11.84TSITDD152 pKa = 3.38VAFSTAWTKK161 pKa = 11.06YY162 pKa = 10.25YY163 pKa = 10.33ILLPDD168 pKa = 4.18PARR171 pKa = 11.84LTMEE175 pKa = 3.78EE176 pKa = 4.28GMFYY180 pKa = 10.59FVDD183 pKa = 3.81TPDD186 pKa = 3.24NGKK189 pKa = 10.73GYY191 pKa = 9.99SFWIDD196 pKa = 3.06EE197 pKa = 4.32LRR199 pKa = 11.84FEE201 pKa = 4.34NSGTIGQVQAYY212 pKa = 7.73IQNGEE217 pKa = 4.35EE218 pKa = 4.07VTSTVFVGQEE228 pKa = 3.99VPVSGLGVSFSLPNGVSQAVNAAPAFFTLASSNPQVAQVVVDD270 pKa = 3.61EE271 pKa = 4.55TTQVSTVRR279 pKa = 11.84ILAQGEE285 pKa = 4.2AVITASMGDD294 pKa = 3.66VAAMGSLTLQVRR306 pKa = 11.84EE307 pKa = 4.59FEE309 pKa = 4.41PAPLPDD315 pKa = 3.99KK316 pKa = 11.06VADD319 pKa = 3.87DD320 pKa = 4.54VISIFSDD327 pKa = 3.34AYY329 pKa = 9.32TNVPVDD335 pKa = 3.97FYY337 pKa = 11.44NGYY340 pKa = 9.09YY341 pKa = 10.63APFQTTVSADD351 pKa = 3.39FTVDD355 pKa = 3.05GDD357 pKa = 4.24HH358 pKa = 6.34VLNYY362 pKa = 10.05INYY365 pKa = 8.29NFVGIEE371 pKa = 3.99FNKK374 pKa = 10.07EE375 pKa = 3.72VPTIDD380 pKa = 3.42ATEE383 pKa = 4.08MTHH386 pKa = 6.19LHH388 pKa = 5.27VDD390 pKa = 3.55IYY392 pKa = 10.7IPEE395 pKa = 4.4ALDD398 pKa = 4.25PNATWRR404 pKa = 11.84VNLRR408 pKa = 11.84DD409 pKa = 4.25FGADD413 pKa = 3.27GNFDD417 pKa = 3.99GGDD420 pKa = 3.53DD421 pKa = 4.27TIASQLLTTASDD433 pKa = 3.64PALVSEE439 pKa = 4.26QWISVDD445 pKa = 2.97MDD447 pKa = 3.04IRR449 pKa = 11.84DD450 pKa = 3.91MANKK454 pKa = 8.38ATLGQIVFDD463 pKa = 4.57AEE465 pKa = 4.79GDD467 pKa = 3.72TSPRR471 pKa = 11.84PSGFYY476 pKa = 10.51VDD478 pKa = 5.44NLYY481 pKa = 10.86LYY483 pKa = 10.38NDD485 pKa = 3.48QGTGGGGPQPIPPTTAAPTPTLDD508 pKa = 3.17AAQVVSIFSDD518 pKa = 3.58AYY520 pKa = 9.95TDD522 pKa = 3.82VPNQGFNQYY531 pKa = 9.31GAATFAEE538 pKa = 4.66IPVADD543 pKa = 4.23NATLNYY549 pKa = 9.04KK550 pKa = 10.35QSDD553 pKa = 3.83NPGGNFQVIEE563 pKa = 4.47LGAEE567 pKa = 3.95NQIDD571 pKa = 3.9ATAMEE576 pKa = 4.64LTDD579 pKa = 3.72FHH581 pKa = 8.03VDD583 pKa = 2.64LWFPNPVNEE592 pKa = 4.07NTAFLLKK599 pKa = 10.24VVNIAPDD606 pKa = 3.33ITSEE610 pKa = 3.79ALIRR614 pKa = 11.84IDD616 pKa = 5.23ANATPAMAQGQWLAIDD632 pKa = 3.94LTMDD636 pKa = 3.3QLQAAGLAANSNIQQVVIDD655 pKa = 4.87LLDD658 pKa = 4.13AGEE661 pKa = 4.27VYY663 pKa = 10.34IDD665 pKa = 3.4NLYY668 pKa = 10.45FSKK671 pKa = 11.26SNGAGGSAPTTSAPEE686 pKa = 3.92PPARR690 pKa = 11.84DD691 pKa = 3.32AADD694 pKa = 3.66VISIFGEE701 pKa = 4.22AYY703 pKa = 9.89TNITGIDD710 pKa = 3.83YY711 pKa = 10.72DD712 pKa = 4.56PNWGQSGHH720 pKa = 5.26MQVNTAFDD728 pKa = 3.68SGDD731 pKa = 3.45GNLALAYY738 pKa = 9.71PSFNYY743 pKa = 10.11QGTDD747 pKa = 3.54FSGNAQNASEE757 pKa = 4.28MEE759 pKa = 4.21FLHH762 pKa = 7.81VDD764 pKa = 3.06IWVPAGTDD772 pKa = 3.22RR773 pKa = 11.84QVKK776 pKa = 9.6VSPINNGTGEE786 pKa = 4.33GEE788 pKa = 4.4VLVSVPLIPGSWNSVDD804 pKa = 5.51LPIADD809 pKa = 3.98FTGMTWDD816 pKa = 3.94SVVQLKK822 pKa = 10.5FDD824 pKa = 3.7GQFNGDD830 pKa = 3.11GSANTDD836 pKa = 3.64PFDD839 pKa = 3.62MYY841 pKa = 11.25LDD843 pKa = 3.25NVYY846 pKa = 10.17FYY848 pKa = 10.44KK849 pKa = 10.75EE850 pKa = 3.86PSAPSAAPTTSAPEE864 pKa = 3.83PLARR868 pKa = 11.84DD869 pKa = 3.38AADD872 pKa = 3.59VISIFGEE879 pKa = 4.22AYY881 pKa = 9.89TNITGIDD888 pKa = 3.83YY889 pKa = 10.72DD890 pKa = 4.56PNWGQSGHH898 pKa = 5.26MQVNTAFDD906 pKa = 4.3PGDD909 pKa = 3.75GNLALAYY916 pKa = 9.71PSFNYY921 pKa = 10.11QGTDD925 pKa = 3.54FSGNAQNASEE935 pKa = 4.28MEE937 pKa = 4.21FLHH940 pKa = 7.81VDD942 pKa = 3.06IWVPAGTDD950 pKa = 3.22RR951 pKa = 11.84QVKK954 pKa = 9.6VSPINNGTGEE964 pKa = 4.33GEE966 pKa = 4.4VLVSVPLTPGAWNSVDD982 pKa = 5.21LPMADD987 pKa = 3.63FTGMTWDD994 pKa = 3.94SVVQLKK1000 pKa = 10.5FDD1002 pKa = 3.7GQFNGDD1008 pKa = 3.11GSANTDD1014 pKa = 3.31PFDD1017 pKa = 3.99IYY1019 pKa = 11.05LDD1021 pKa = 3.23NVYY1024 pKa = 10.14FYY1026 pKa = 10.44KK1027 pKa = 10.7EE1028 pKa = 3.75PSAPSTAPTTSAPAPPVRR1046 pKa = 11.84DD1047 pKa = 3.33AADD1050 pKa = 3.61VISIFGEE1057 pKa = 4.22AYY1059 pKa = 9.89TNITGIDD1066 pKa = 3.83YY1067 pKa = 10.72DD1068 pKa = 4.56PNWGQSGHH1076 pKa = 5.26MQVNTAFDD1084 pKa = 4.3PGDD1087 pKa = 3.75GNLALAYY1094 pKa = 9.71PSFNYY1099 pKa = 10.11QGTDD1103 pKa = 3.54FSGNAQNAATMQFLHH1118 pKa = 7.2VDD1120 pKa = 3.2IWVPAGTDD1128 pKa = 3.22RR1129 pKa = 11.84QVKK1132 pKa = 9.6VSPINNGTGAGEE1144 pKa = 4.21VLVSVPLTPGAWNSVDD1160 pKa = 4.95LPIGDD1165 pKa = 4.06FTGMTWDD1172 pKa = 4.28SVFQLKK1178 pKa = 10.32FDD1180 pKa = 3.85GQFNGDD1186 pKa = 3.11GSANTDD1192 pKa = 3.31PFDD1195 pKa = 3.99IYY1197 pKa = 11.05LDD1199 pKa = 3.23NVYY1202 pKa = 10.14FYY1204 pKa = 10.44KK1205 pKa = 10.7EE1206 pKa = 3.75PSAPSTAPTTSAPAPPVRR1224 pKa = 11.84DD1225 pKa = 3.33AADD1228 pKa = 3.61VISIFGEE1235 pKa = 4.22AYY1237 pKa = 9.67TNITGVDD1244 pKa = 3.77YY1245 pKa = 11.05DD1246 pKa = 4.63PNWGQSGHH1254 pKa = 5.26MQVNTAFDD1262 pKa = 4.3PGDD1265 pKa = 3.75GNLALAYY1272 pKa = 9.89PNFNYY1277 pKa = 10.28QGTDD1281 pKa = 3.53FSGNAQNAASMQFLHH1296 pKa = 7.39VDD1298 pKa = 3.07IWVPAGTDD1306 pKa = 3.22RR1307 pKa = 11.84QVKK1310 pKa = 9.6VSPINNGTGAGEE1322 pKa = 4.21VLVSVPLTPGAWNSVDD1338 pKa = 4.95LPIGDD1343 pKa = 4.06FTGMTWDD1350 pKa = 4.28SVFQLKK1356 pKa = 10.32FDD1358 pKa = 3.85GQFNGDD1364 pKa = 3.11GSANTDD1370 pKa = 3.42PFDD1373 pKa = 3.73VYY1375 pKa = 11.29LDD1377 pKa = 3.31NVYY1380 pKa = 10.22FYY1382 pKa = 11.57KK1383 pKa = 10.9NN1384 pKa = 3.04
Molecular weight: 148.64 kDa
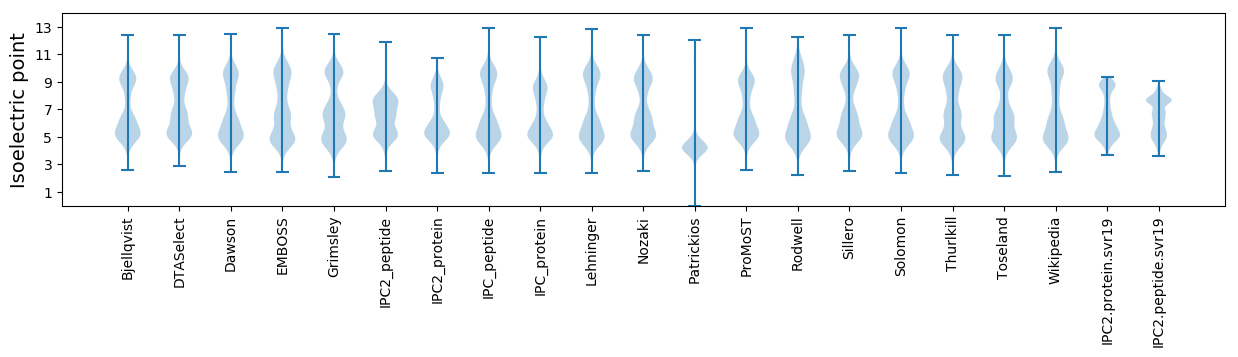
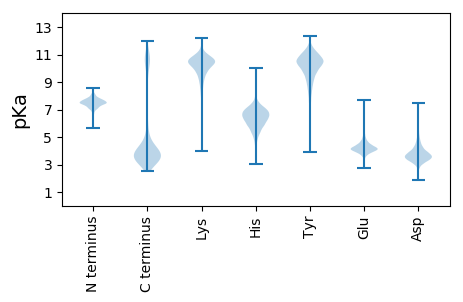
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A4G414|A0A2A4G414_9FLAO Uncharacterized protein OS=Sediminicola luteus OX=319238 GN=B7P33_15560 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 6.16KK51 pKa = 10.53KK52 pKa = 10.04
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 6.16KK51 pKa = 10.53KK52 pKa = 10.04
Molecular weight: 6.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1364760 |
31 |
6537 |
361.9 |
40.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.441 ± 0.054 | 0.756 ± 0.015 |
5.85 ± 0.049 | 6.286 ± 0.043 |
4.905 ± 0.029 | 7.306 ± 0.04 |
2.093 ± 0.021 | 6.441 ± 0.036 |
6.523 ± 0.06 | 9.711 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.387 ± 0.024 | 5.083 ± 0.047 |
3.978 ± 0.026 | 3.978 ± 0.025 |
3.981 ± 0.034 | 5.952 ± 0.031 |
5.814 ± 0.064 | 6.313 ± 0.032 |
1.261 ± 0.014 | 3.941 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
