
Apis mellifera associated microvirus 32
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.65
Get precalculated fractions of proteins
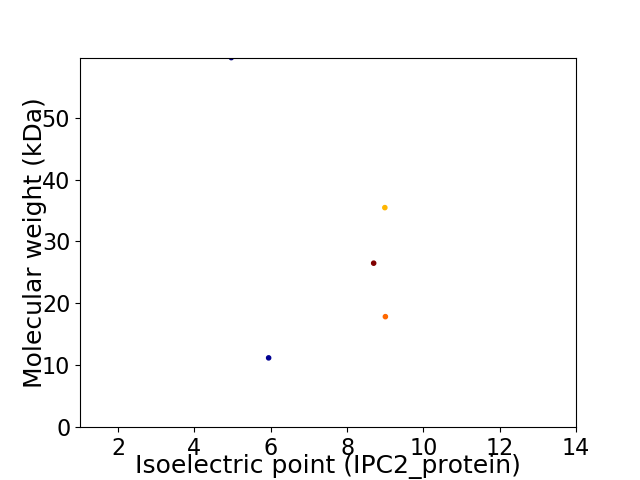
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTX9|A0A3S8UTX9_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 32 OX=2494761 PE=3 SV=1
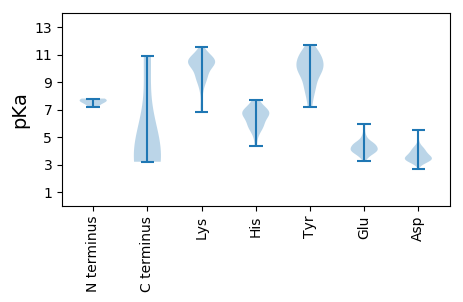
MM1 pKa = 7.57GFRR4 pKa = 11.84VNTAGRR10 pKa = 11.84VKK12 pKa = 10.23QSHH15 pKa = 6.79FSQVPANVAAPRR27 pKa = 11.84SPFDD31 pKa = 3.45RR32 pKa = 11.84SFSHH36 pKa = 6.23KK37 pKa = 7.25TTINEE42 pKa = 3.77GFLYY46 pKa = 9.99PIFWEE51 pKa = 5.08PILPGDD57 pKa = 4.02TVNLTMNCLARR68 pKa = 11.84LATPIFPYY76 pKa = 9.66MDD78 pKa = 3.37NVYY81 pKa = 10.93LDD83 pKa = 3.59VHH85 pKa = 6.11WFFVPNRR92 pKa = 11.84LVWNHH97 pKa = 5.56WEE99 pKa = 4.07QFQGAQDD106 pKa = 3.94NPPDD110 pKa = 3.6TFTDD114 pKa = 3.92YY115 pKa = 10.85EE116 pKa = 4.6VPSLDD121 pKa = 4.2DD122 pKa = 3.38ATHH125 pKa = 6.71AGGFASGSLYY135 pKa = 10.54DD136 pKa = 4.16YY137 pKa = 10.87FGLPTLVTGIDD148 pKa = 3.63QTNMPIALPFRR159 pKa = 11.84AYY161 pKa = 10.5RR162 pKa = 11.84KK163 pKa = 8.99IWNEE167 pKa = 3.41WYY169 pKa = 10.12RR170 pKa = 11.84DD171 pKa = 3.84EE172 pKa = 5.62NSQDD176 pKa = 3.59PLTVDD181 pKa = 3.52MGDD184 pKa = 4.06GPDD187 pKa = 3.41TTDD190 pKa = 3.08YY191 pKa = 11.72SLLKK195 pKa = 10.22RR196 pKa = 11.84NRR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 10.66DD201 pKa = 3.53YY202 pKa = 8.17FTSCLPWPQKK212 pKa = 10.78GPAVTIPIGSGSLPVVLNPDD232 pKa = 5.51AIPDD236 pKa = 3.61SPIIRR241 pKa = 11.84RR242 pKa = 11.84ADD244 pKa = 3.49NQTSSQNSLTGLITSGAGALRR265 pKa = 11.84GSDD268 pKa = 3.42SFNSVIDD275 pKa = 4.14PNDD278 pKa = 3.42TLMVEE283 pKa = 4.59LDD285 pKa = 3.53DD286 pKa = 4.33VAAVSINTLRR296 pKa = 11.84EE297 pKa = 4.3SIVLQQMLEE306 pKa = 3.72LDD308 pKa = 3.49ARR310 pKa = 11.84GGTRR314 pKa = 11.84YY315 pKa = 9.91VEE317 pKa = 4.03ALLARR322 pKa = 11.84FGVVSPDD329 pKa = 3.21FRR331 pKa = 11.84LQRR334 pKa = 11.84PEE336 pKa = 3.8FLGGQTFDD344 pKa = 3.95INVSPIAQTSSTDD357 pKa = 3.1AEE359 pKa = 4.67TPQGNLSGFAVARR372 pKa = 11.84GSSQLSHH379 pKa = 7.06SFVEE383 pKa = 4.55HH384 pKa = 5.93GQLLGLVSVRR394 pKa = 11.84ADD396 pKa = 3.01ATYY399 pKa = 8.72QQGMSRR405 pKa = 11.84HH406 pKa = 4.32WSVRR410 pKa = 11.84TRR412 pKa = 11.84YY413 pKa = 10.14DD414 pKa = 3.16YY415 pKa = 11.52YY416 pKa = 11.22EE417 pKa = 4.36PLAANLGEE425 pKa = 3.93QAVLNKK431 pKa = 9.55EE432 pKa = 3.81IYY434 pKa = 7.36MTGTEE439 pKa = 4.1ATDD442 pKa = 3.38NAVFGYY448 pKa = 7.17QEE450 pKa = 3.24RR451 pKa = 11.84WAEE454 pKa = 3.85YY455 pKa = 9.34RR456 pKa = 11.84YY457 pKa = 10.22KK458 pKa = 10.43PSYY461 pKa = 7.85VTGIFRR467 pKa = 11.84SNAAASLDD475 pKa = 3.73SWHH478 pKa = 6.71LAIEE482 pKa = 4.76FGSLPTLADD491 pKa = 4.78LLPEE495 pKa = 4.45SPPIDD500 pKa = 4.15RR501 pKa = 11.84IVAVPSEE508 pKa = 3.85PHH510 pKa = 6.76FIVDD514 pKa = 3.15TWTKK518 pKa = 8.78FRR520 pKa = 11.84HH521 pKa = 4.95VRR523 pKa = 11.84VLPVYY528 pKa = 9.77SAPGLTRR535 pKa = 11.84LL536 pKa = 4.24
MM1 pKa = 7.57GFRR4 pKa = 11.84VNTAGRR10 pKa = 11.84VKK12 pKa = 10.23QSHH15 pKa = 6.79FSQVPANVAAPRR27 pKa = 11.84SPFDD31 pKa = 3.45RR32 pKa = 11.84SFSHH36 pKa = 6.23KK37 pKa = 7.25TTINEE42 pKa = 3.77GFLYY46 pKa = 9.99PIFWEE51 pKa = 5.08PILPGDD57 pKa = 4.02TVNLTMNCLARR68 pKa = 11.84LATPIFPYY76 pKa = 9.66MDD78 pKa = 3.37NVYY81 pKa = 10.93LDD83 pKa = 3.59VHH85 pKa = 6.11WFFVPNRR92 pKa = 11.84LVWNHH97 pKa = 5.56WEE99 pKa = 4.07QFQGAQDD106 pKa = 3.94NPPDD110 pKa = 3.6TFTDD114 pKa = 3.92YY115 pKa = 10.85EE116 pKa = 4.6VPSLDD121 pKa = 4.2DD122 pKa = 3.38ATHH125 pKa = 6.71AGGFASGSLYY135 pKa = 10.54DD136 pKa = 4.16YY137 pKa = 10.87FGLPTLVTGIDD148 pKa = 3.63QTNMPIALPFRR159 pKa = 11.84AYY161 pKa = 10.5RR162 pKa = 11.84KK163 pKa = 8.99IWNEE167 pKa = 3.41WYY169 pKa = 10.12RR170 pKa = 11.84DD171 pKa = 3.84EE172 pKa = 5.62NSQDD176 pKa = 3.59PLTVDD181 pKa = 3.52MGDD184 pKa = 4.06GPDD187 pKa = 3.41TTDD190 pKa = 3.08YY191 pKa = 11.72SLLKK195 pKa = 10.22RR196 pKa = 11.84NRR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 10.66DD201 pKa = 3.53YY202 pKa = 8.17FTSCLPWPQKK212 pKa = 10.78GPAVTIPIGSGSLPVVLNPDD232 pKa = 5.51AIPDD236 pKa = 3.61SPIIRR241 pKa = 11.84RR242 pKa = 11.84ADD244 pKa = 3.49NQTSSQNSLTGLITSGAGALRR265 pKa = 11.84GSDD268 pKa = 3.42SFNSVIDD275 pKa = 4.14PNDD278 pKa = 3.42TLMVEE283 pKa = 4.59LDD285 pKa = 3.53DD286 pKa = 4.33VAAVSINTLRR296 pKa = 11.84EE297 pKa = 4.3SIVLQQMLEE306 pKa = 3.72LDD308 pKa = 3.49ARR310 pKa = 11.84GGTRR314 pKa = 11.84YY315 pKa = 9.91VEE317 pKa = 4.03ALLARR322 pKa = 11.84FGVVSPDD329 pKa = 3.21FRR331 pKa = 11.84LQRR334 pKa = 11.84PEE336 pKa = 3.8FLGGQTFDD344 pKa = 3.95INVSPIAQTSSTDD357 pKa = 3.1AEE359 pKa = 4.67TPQGNLSGFAVARR372 pKa = 11.84GSSQLSHH379 pKa = 7.06SFVEE383 pKa = 4.55HH384 pKa = 5.93GQLLGLVSVRR394 pKa = 11.84ADD396 pKa = 3.01ATYY399 pKa = 8.72QQGMSRR405 pKa = 11.84HH406 pKa = 4.32WSVRR410 pKa = 11.84TRR412 pKa = 11.84YY413 pKa = 10.14DD414 pKa = 3.16YY415 pKa = 11.52YY416 pKa = 11.22EE417 pKa = 4.36PLAANLGEE425 pKa = 3.93QAVLNKK431 pKa = 9.55EE432 pKa = 3.81IYY434 pKa = 7.36MTGTEE439 pKa = 4.1ATDD442 pKa = 3.38NAVFGYY448 pKa = 7.17QEE450 pKa = 3.24RR451 pKa = 11.84WAEE454 pKa = 3.85YY455 pKa = 9.34RR456 pKa = 11.84YY457 pKa = 10.22KK458 pKa = 10.43PSYY461 pKa = 7.85VTGIFRR467 pKa = 11.84SNAAASLDD475 pKa = 3.73SWHH478 pKa = 6.71LAIEE482 pKa = 4.76FGSLPTLADD491 pKa = 4.78LLPEE495 pKa = 4.45SPPIDD500 pKa = 4.15RR501 pKa = 11.84IVAVPSEE508 pKa = 3.85PHH510 pKa = 6.76FIVDD514 pKa = 3.15TWTKK518 pKa = 8.78FRR520 pKa = 11.84HH521 pKa = 4.95VRR523 pKa = 11.84VLPVYY528 pKa = 9.77SAPGLTRR535 pKa = 11.84LL536 pKa = 4.24
Molecular weight: 59.69 kDa
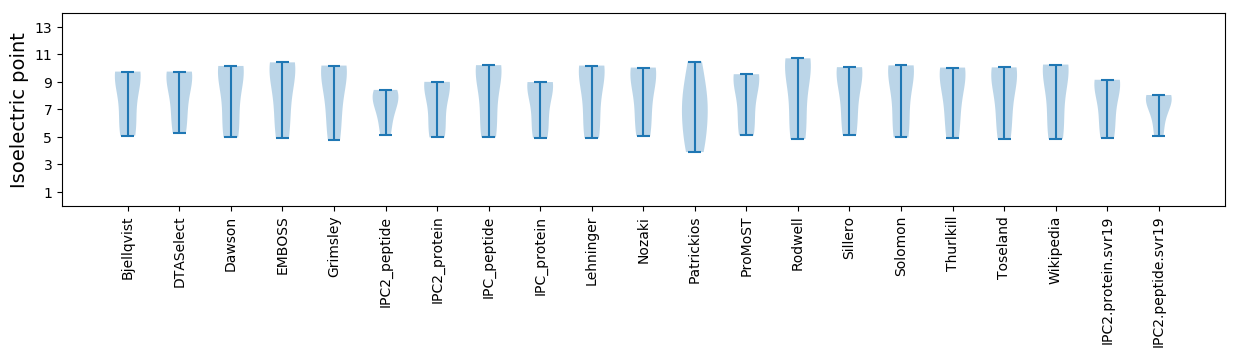
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTX9|A0A3S8UTX9_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 32 OX=2494761 PE=3 SV=1
MM1 pKa = 7.8PCYY4 pKa = 10.14KK5 pKa = 9.79PLKK8 pKa = 10.47AMVTRR13 pKa = 11.84SPSGGKK19 pKa = 8.08STVRR23 pKa = 11.84FHH25 pKa = 7.49KK26 pKa = 10.37GLKK29 pKa = 8.54LTACEE34 pKa = 4.36AKK36 pKa = 10.33HH37 pKa = 5.33GTATPIPCGQCVGCRR52 pKa = 11.84LEE54 pKa = 5.96RR55 pKa = 11.84SRR57 pKa = 11.84QWAIRR62 pKa = 11.84LMKK65 pKa = 9.83EE66 pKa = 4.05LKK68 pKa = 10.52LHH70 pKa = 6.53DD71 pKa = 4.08RR72 pKa = 11.84SSFLTLTYY80 pKa = 11.03DD81 pKa = 3.82DD82 pKa = 3.86AHH84 pKa = 6.94LPRR87 pKa = 11.84LRR89 pKa = 11.84NGLPTLVLEE98 pKa = 5.86DD99 pKa = 3.06IQLFLKK105 pKa = 10.58RR106 pKa = 11.84LRR108 pKa = 11.84KK109 pKa = 9.23EE110 pKa = 3.52LALPPEE116 pKa = 4.22SLRR119 pKa = 11.84YY120 pKa = 8.65FQAGEE125 pKa = 4.09YY126 pKa = 11.06GEE128 pKa = 4.54VTHH131 pKa = 6.97RR132 pKa = 11.84PHH134 pKa = 7.53HH135 pKa = 5.82HH136 pKa = 6.54MILFGEE142 pKa = 4.45DD143 pKa = 3.48FSRR146 pKa = 11.84DD147 pKa = 3.29RR148 pKa = 11.84VRR150 pKa = 11.84VADD153 pKa = 3.71SRR155 pKa = 11.84SGYY158 pKa = 10.65AQFEE162 pKa = 4.77SEE164 pKa = 5.12LLTSLWGKK172 pKa = 8.96GRR174 pKa = 11.84CTISEE179 pKa = 4.18VSFEE183 pKa = 4.05SAAYY187 pKa = 8.35VARR190 pKa = 11.84YY191 pKa = 8.74CLKK194 pKa = 10.36KK195 pKa = 9.87VTGKK199 pKa = 10.65GSRR202 pKa = 11.84FHH204 pKa = 5.93YY205 pKa = 9.94RR206 pKa = 11.84GRR208 pKa = 11.84KK209 pKa = 8.27PEE211 pKa = 3.95FVTMSRR217 pKa = 11.84RR218 pKa = 11.84PGIAARR224 pKa = 11.84YY225 pKa = 8.68FEE227 pKa = 4.61EE228 pKa = 4.82FKK230 pKa = 11.21SDD232 pKa = 3.4IYY234 pKa = 10.95PHH236 pKa = 7.15DD237 pKa = 3.92EE238 pKa = 4.13VVPGPGRR245 pKa = 11.84PASLPPKK252 pKa = 10.42YY253 pKa = 9.85FDD255 pKa = 4.2KK256 pKa = 11.06LLEE259 pKa = 4.43KK260 pKa = 10.49VDD262 pKa = 3.76PALFAEE268 pKa = 4.62IKK270 pKa = 9.53KK271 pKa = 10.49KK272 pKa = 10.42RR273 pKa = 11.84VEE275 pKa = 4.95GLDD278 pKa = 3.39FWSDD282 pKa = 3.52PNSTDD287 pKa = 2.67TRR289 pKa = 11.84LEE291 pKa = 3.55TRR293 pKa = 11.84EE294 pKa = 3.9RR295 pKa = 11.84VKK297 pKa = 10.98EE298 pKa = 3.96SLIKK302 pKa = 10.47NCLKK306 pKa = 10.78RR307 pKa = 11.84SVV309 pKa = 3.2
MM1 pKa = 7.8PCYY4 pKa = 10.14KK5 pKa = 9.79PLKK8 pKa = 10.47AMVTRR13 pKa = 11.84SPSGGKK19 pKa = 8.08STVRR23 pKa = 11.84FHH25 pKa = 7.49KK26 pKa = 10.37GLKK29 pKa = 8.54LTACEE34 pKa = 4.36AKK36 pKa = 10.33HH37 pKa = 5.33GTATPIPCGQCVGCRR52 pKa = 11.84LEE54 pKa = 5.96RR55 pKa = 11.84SRR57 pKa = 11.84QWAIRR62 pKa = 11.84LMKK65 pKa = 9.83EE66 pKa = 4.05LKK68 pKa = 10.52LHH70 pKa = 6.53DD71 pKa = 4.08RR72 pKa = 11.84SSFLTLTYY80 pKa = 11.03DD81 pKa = 3.82DD82 pKa = 3.86AHH84 pKa = 6.94LPRR87 pKa = 11.84LRR89 pKa = 11.84NGLPTLVLEE98 pKa = 5.86DD99 pKa = 3.06IQLFLKK105 pKa = 10.58RR106 pKa = 11.84LRR108 pKa = 11.84KK109 pKa = 9.23EE110 pKa = 3.52LALPPEE116 pKa = 4.22SLRR119 pKa = 11.84YY120 pKa = 8.65FQAGEE125 pKa = 4.09YY126 pKa = 11.06GEE128 pKa = 4.54VTHH131 pKa = 6.97RR132 pKa = 11.84PHH134 pKa = 7.53HH135 pKa = 5.82HH136 pKa = 6.54MILFGEE142 pKa = 4.45DD143 pKa = 3.48FSRR146 pKa = 11.84DD147 pKa = 3.29RR148 pKa = 11.84VRR150 pKa = 11.84VADD153 pKa = 3.71SRR155 pKa = 11.84SGYY158 pKa = 10.65AQFEE162 pKa = 4.77SEE164 pKa = 5.12LLTSLWGKK172 pKa = 8.96GRR174 pKa = 11.84CTISEE179 pKa = 4.18VSFEE183 pKa = 4.05SAAYY187 pKa = 8.35VARR190 pKa = 11.84YY191 pKa = 8.74CLKK194 pKa = 10.36KK195 pKa = 9.87VTGKK199 pKa = 10.65GSRR202 pKa = 11.84FHH204 pKa = 5.93YY205 pKa = 9.94RR206 pKa = 11.84GRR208 pKa = 11.84KK209 pKa = 8.27PEE211 pKa = 3.95FVTMSRR217 pKa = 11.84RR218 pKa = 11.84PGIAARR224 pKa = 11.84YY225 pKa = 8.68FEE227 pKa = 4.61EE228 pKa = 4.82FKK230 pKa = 11.21SDD232 pKa = 3.4IYY234 pKa = 10.95PHH236 pKa = 7.15DD237 pKa = 3.92EE238 pKa = 4.13VVPGPGRR245 pKa = 11.84PASLPPKK252 pKa = 10.42YY253 pKa = 9.85FDD255 pKa = 4.2KK256 pKa = 11.06LLEE259 pKa = 4.43KK260 pKa = 10.49VDD262 pKa = 3.76PALFAEE268 pKa = 4.62IKK270 pKa = 9.53KK271 pKa = 10.49KK272 pKa = 10.42RR273 pKa = 11.84VEE275 pKa = 4.95GLDD278 pKa = 3.39FWSDD282 pKa = 3.52PNSTDD287 pKa = 2.67TRR289 pKa = 11.84LEE291 pKa = 3.55TRR293 pKa = 11.84EE294 pKa = 3.9RR295 pKa = 11.84VKK297 pKa = 10.98EE298 pKa = 3.96SLIKK302 pKa = 10.47NCLKK306 pKa = 10.78RR307 pKa = 11.84SVV309 pKa = 3.2
Molecular weight: 35.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1361 |
102 |
536 |
272.2 |
30.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.405 ± 1.88 | 0.808 ± 0.518 |
5.511 ± 0.609 | 5.731 ± 0.745 |
4.482 ± 0.926 | 6.98 ± 0.441 |
2.278 ± 0.315 | 3.6 ± 0.382 |
4.923 ± 1.42 | 8.891 ± 0.745 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.278 ± 0.492 | 3.821 ± 0.873 |
6.466 ± 1.117 | 3.527 ± 0.554 |
7.274 ± 0.803 | 8.597 ± 0.706 |
5.364 ± 0.792 | 6.025 ± 0.569 |
1.176 ± 0.416 | 2.866 ± 0.688 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
