
Collinsella stercoris DSM 13279
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; Collinsella stercoris
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
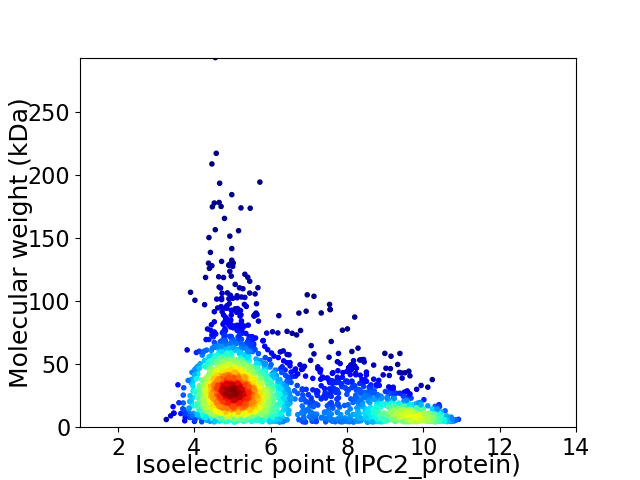
Virtual 2D-PAGE plot for 2527 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B6GA64|B6GA64_9ACTN Toxin-antitoxin system antitoxin component HicB family OS=Collinsella stercoris DSM 13279 OX=445975 GN=COLSTE_00959 PE=4 SV=1
MM1 pKa = 7.09PQHH4 pKa = 6.6RR5 pKa = 11.84NKK7 pKa = 10.82AVVAGLAVTLAFGGVALPAVASTEE31 pKa = 3.99QAPATDD37 pKa = 3.85SDD39 pKa = 4.56PNGVGDD45 pKa = 4.85PAPADD50 pKa = 3.69YY51 pKa = 11.2DD52 pKa = 3.5KK53 pKa = 11.66ADD55 pKa = 3.77YY56 pKa = 11.4YY57 pKa = 11.14EE58 pKa = 4.4GTEE61 pKa = 4.3GVATFASARR70 pKa = 11.84SASTLSPVSLSDD82 pKa = 3.13EE83 pKa = 3.92MKK85 pKa = 10.77YY86 pKa = 8.24FTKK89 pKa = 10.87YY90 pKa = 10.74EE91 pKa = 4.17SGCDD95 pKa = 3.54YY96 pKa = 11.44DD97 pKa = 5.28KK98 pKa = 11.54GLSYY102 pKa = 11.55GDD104 pKa = 4.54GYY106 pKa = 11.64NAMGYY111 pKa = 8.13YY112 pKa = 9.84QFDD115 pKa = 3.2RR116 pKa = 11.84RR117 pKa = 11.84YY118 pKa = 9.89SLLPFIEE125 pKa = 4.0QVYY128 pKa = 10.17GYY130 pKa = 10.84NSTKK134 pKa = 10.53YY135 pKa = 11.11DD136 pKa = 3.51MLRR139 pKa = 11.84AALAYY144 pKa = 10.1RR145 pKa = 11.84DD146 pKa = 3.85VLQNPSYY153 pKa = 11.37EE154 pKa = 4.27MYY156 pKa = 10.36DD157 pKa = 3.38YY158 pKa = 11.11TSRR161 pKa = 11.84QLSAPAQILNGAWHH175 pKa = 7.16AAYY178 pKa = 9.87AADD181 pKa = 3.84PAEE184 pKa = 4.75FSALQDD190 pKa = 3.16SYY192 pKa = 12.07AYY194 pKa = 10.53NNYY197 pKa = 8.97YY198 pKa = 10.8LPVQSSLLNTYY209 pKa = 10.16GVDD212 pKa = 3.22VRR214 pKa = 11.84DD215 pKa = 4.05RR216 pKa = 11.84ADD218 pKa = 3.5CVKK221 pKa = 10.6GLVWGMCNLFGQGGVQKK238 pKa = 10.17FFKK241 pKa = 10.4GANIDD246 pKa = 3.2NSMTDD251 pKa = 3.18RR252 pKa = 11.84EE253 pKa = 5.27LITALCDD260 pKa = 3.54TVVEE264 pKa = 4.92YY265 pKa = 11.38VDD267 pKa = 4.42DD268 pKa = 4.82WYY270 pKa = 10.38PSQPQYY276 pKa = 10.04WDD278 pKa = 2.85GWKK281 pKa = 10.03NRR283 pKa = 11.84YY284 pKa = 9.14KK285 pKa = 10.52KK286 pKa = 10.68EE287 pKa = 3.84KK288 pKa = 8.66ATCLAYY294 pKa = 9.64MDD296 pKa = 4.05QHH298 pKa = 7.08DD299 pKa = 4.75AEE301 pKa = 4.55QNANGQGQTDD311 pKa = 3.93DD312 pKa = 4.05QPNADD317 pKa = 4.82EE318 pKa = 5.23PGQDD322 pKa = 3.87DD323 pKa = 4.2SGAGLPIAPDD333 pKa = 3.44APNGSEE339 pKa = 4.01GQNGDD344 pKa = 3.32GGGSQGDD351 pKa = 3.7ADD353 pKa = 4.1SGEE356 pKa = 4.3SGSDD360 pKa = 2.68GGAAHH365 pKa = 6.97GGDD368 pKa = 3.93SGSDD372 pKa = 3.35GAVTPEE378 pKa = 4.06GGQGSDD384 pKa = 3.22AEE386 pKa = 4.31QNAGGSQNSDD396 pKa = 3.79DD397 pKa = 5.28DD398 pKa = 6.26ADD400 pKa = 4.19DD401 pKa = 4.47APNASVPDD409 pKa = 4.13GNAGQGSSGDD419 pKa = 3.54QEE421 pKa = 4.26NAGGSGSGSAGNSGSAQGGTGNNGGSTQDD450 pKa = 3.35GSAGTGNSAGTDD462 pKa = 3.24GSTGNGSTQDD472 pKa = 3.28TTTGGGSTHH481 pKa = 6.84AGEE484 pKa = 4.84GQGSSDD490 pKa = 5.43DD491 pKa = 4.07EE492 pKa = 4.26QTDD495 pKa = 3.14ASTGDD500 pKa = 3.66EE501 pKa = 4.13EE502 pKa = 6.54DD503 pKa = 4.89DD504 pKa = 4.67GKK506 pKa = 8.19TTPKK510 pKa = 10.82DD511 pKa = 3.15PAPQQNTTRR520 pKa = 11.84DD521 pKa = 3.61PMNSLGTTGDD531 pKa = 3.48KK532 pKa = 7.71TTSGQGDD539 pKa = 4.0GAQGTSASGAGKK551 pKa = 8.6ATAGGLPGTGDD562 pKa = 3.34IATMVLTGAAGLAVAGSSFLSLAKK586 pKa = 10.15KK587 pKa = 9.67EE588 pKa = 4.0RR589 pKa = 11.84AGVDD593 pKa = 3.54EE594 pKa = 4.83SADD597 pKa = 3.9SNDD600 pKa = 3.36EE601 pKa = 3.96
MM1 pKa = 7.09PQHH4 pKa = 6.6RR5 pKa = 11.84NKK7 pKa = 10.82AVVAGLAVTLAFGGVALPAVASTEE31 pKa = 3.99QAPATDD37 pKa = 3.85SDD39 pKa = 4.56PNGVGDD45 pKa = 4.85PAPADD50 pKa = 3.69YY51 pKa = 11.2DD52 pKa = 3.5KK53 pKa = 11.66ADD55 pKa = 3.77YY56 pKa = 11.4YY57 pKa = 11.14EE58 pKa = 4.4GTEE61 pKa = 4.3GVATFASARR70 pKa = 11.84SASTLSPVSLSDD82 pKa = 3.13EE83 pKa = 3.92MKK85 pKa = 10.77YY86 pKa = 8.24FTKK89 pKa = 10.87YY90 pKa = 10.74EE91 pKa = 4.17SGCDD95 pKa = 3.54YY96 pKa = 11.44DD97 pKa = 5.28KK98 pKa = 11.54GLSYY102 pKa = 11.55GDD104 pKa = 4.54GYY106 pKa = 11.64NAMGYY111 pKa = 8.13YY112 pKa = 9.84QFDD115 pKa = 3.2RR116 pKa = 11.84RR117 pKa = 11.84YY118 pKa = 9.89SLLPFIEE125 pKa = 4.0QVYY128 pKa = 10.17GYY130 pKa = 10.84NSTKK134 pKa = 10.53YY135 pKa = 11.11DD136 pKa = 3.51MLRR139 pKa = 11.84AALAYY144 pKa = 10.1RR145 pKa = 11.84DD146 pKa = 3.85VLQNPSYY153 pKa = 11.37EE154 pKa = 4.27MYY156 pKa = 10.36DD157 pKa = 3.38YY158 pKa = 11.11TSRR161 pKa = 11.84QLSAPAQILNGAWHH175 pKa = 7.16AAYY178 pKa = 9.87AADD181 pKa = 3.84PAEE184 pKa = 4.75FSALQDD190 pKa = 3.16SYY192 pKa = 12.07AYY194 pKa = 10.53NNYY197 pKa = 8.97YY198 pKa = 10.8LPVQSSLLNTYY209 pKa = 10.16GVDD212 pKa = 3.22VRR214 pKa = 11.84DD215 pKa = 4.05RR216 pKa = 11.84ADD218 pKa = 3.5CVKK221 pKa = 10.6GLVWGMCNLFGQGGVQKK238 pKa = 10.17FFKK241 pKa = 10.4GANIDD246 pKa = 3.2NSMTDD251 pKa = 3.18RR252 pKa = 11.84EE253 pKa = 5.27LITALCDD260 pKa = 3.54TVVEE264 pKa = 4.92YY265 pKa = 11.38VDD267 pKa = 4.42DD268 pKa = 4.82WYY270 pKa = 10.38PSQPQYY276 pKa = 10.04WDD278 pKa = 2.85GWKK281 pKa = 10.03NRR283 pKa = 11.84YY284 pKa = 9.14KK285 pKa = 10.52KK286 pKa = 10.68EE287 pKa = 3.84KK288 pKa = 8.66ATCLAYY294 pKa = 9.64MDD296 pKa = 4.05QHH298 pKa = 7.08DD299 pKa = 4.75AEE301 pKa = 4.55QNANGQGQTDD311 pKa = 3.93DD312 pKa = 4.05QPNADD317 pKa = 4.82EE318 pKa = 5.23PGQDD322 pKa = 3.87DD323 pKa = 4.2SGAGLPIAPDD333 pKa = 3.44APNGSEE339 pKa = 4.01GQNGDD344 pKa = 3.32GGGSQGDD351 pKa = 3.7ADD353 pKa = 4.1SGEE356 pKa = 4.3SGSDD360 pKa = 2.68GGAAHH365 pKa = 6.97GGDD368 pKa = 3.93SGSDD372 pKa = 3.35GAVTPEE378 pKa = 4.06GGQGSDD384 pKa = 3.22AEE386 pKa = 4.31QNAGGSQNSDD396 pKa = 3.79DD397 pKa = 5.28DD398 pKa = 6.26ADD400 pKa = 4.19DD401 pKa = 4.47APNASVPDD409 pKa = 4.13GNAGQGSSGDD419 pKa = 3.54QEE421 pKa = 4.26NAGGSGSGSAGNSGSAQGGTGNNGGSTQDD450 pKa = 3.35GSAGTGNSAGTDD462 pKa = 3.24GSTGNGSTQDD472 pKa = 3.28TTTGGGSTHH481 pKa = 6.84AGEE484 pKa = 4.84GQGSSDD490 pKa = 5.43DD491 pKa = 4.07EE492 pKa = 4.26QTDD495 pKa = 3.14ASTGDD500 pKa = 3.66EE501 pKa = 4.13EE502 pKa = 6.54DD503 pKa = 4.89DD504 pKa = 4.67GKK506 pKa = 8.19TTPKK510 pKa = 10.82DD511 pKa = 3.15PAPQQNTTRR520 pKa = 11.84DD521 pKa = 3.61PMNSLGTTGDD531 pKa = 3.48KK532 pKa = 7.71TTSGQGDD539 pKa = 4.0GAQGTSASGAGKK551 pKa = 8.6ATAGGLPGTGDD562 pKa = 3.34IATMVLTGAAGLAVAGSSFLSLAKK586 pKa = 10.15KK587 pKa = 9.67EE588 pKa = 4.0RR589 pKa = 11.84AGVDD593 pKa = 3.54EE594 pKa = 4.83SADD597 pKa = 3.9SNDD600 pKa = 3.36EE601 pKa = 3.96
Molecular weight: 61.33 kDa
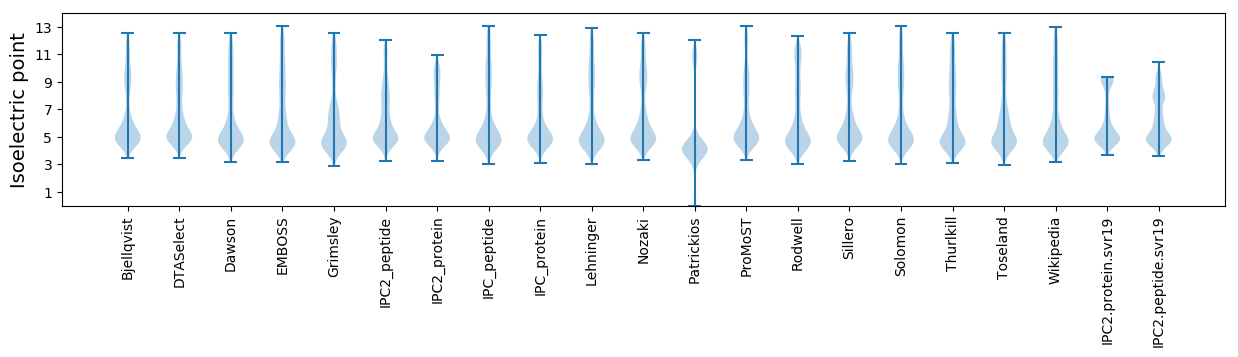
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B6GD79|B6GD79_9ACTN Serine-type D-Ala-D-Ala carboxypeptidase OS=Collinsella stercoris DSM 13279 OX=445975 GN=COLSTE_02057 PE=3 SV=1
MM1 pKa = 7.52HH2 pKa = 7.32GGAARR7 pKa = 11.84PIGLGVGARR16 pKa = 11.84AFSPKK21 pKa = 9.83RR22 pKa = 11.84PRR24 pKa = 11.84LHH26 pKa = 6.76AWRR29 pKa = 11.84PCAGVFFSFLGRR41 pKa = 11.84GRR43 pKa = 11.84SIRR46 pKa = 11.84VRR48 pKa = 3.4
MM1 pKa = 7.52HH2 pKa = 7.32GGAARR7 pKa = 11.84PIGLGVGARR16 pKa = 11.84AFSPKK21 pKa = 9.83RR22 pKa = 11.84PRR24 pKa = 11.84LHH26 pKa = 6.76AWRR29 pKa = 11.84PCAGVFFSFLGRR41 pKa = 11.84GRR43 pKa = 11.84SIRR46 pKa = 11.84VRR48 pKa = 3.4
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
703773 |
39 |
2718 |
278.5 |
30.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.725 ± 0.075 | 1.59 ± 0.021 |
6.19 ± 0.045 | 6.598 ± 0.051 |
3.612 ± 0.036 | 8.324 ± 0.045 |
2.022 ± 0.024 | 5.025 ± 0.04 |
3.546 ± 0.046 | 9.099 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.77 ± 0.028 | 2.872 ± 0.034 |
4.439 ± 0.036 | 2.849 ± 0.032 |
6.566 ± 0.065 | 5.849 ± 0.042 |
5.269 ± 0.041 | 7.826 ± 0.05 |
1.095 ± 0.02 | 2.736 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
