
Marvinbryantia formatexigens DSM 14469
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Marvinbryantia; Marvinbryantia formatexigens
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
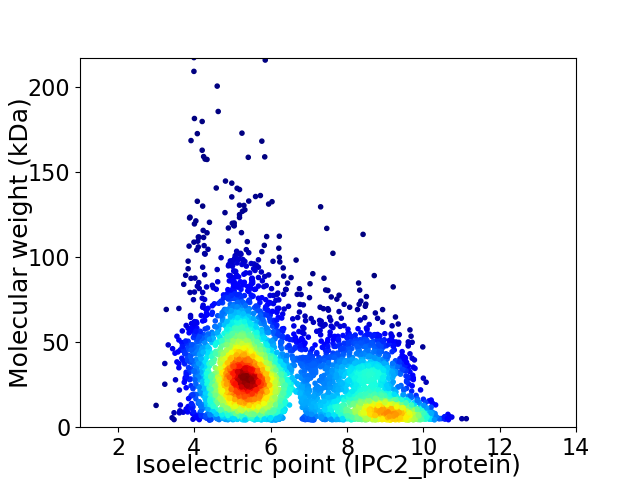
Virtual 2D-PAGE plot for 4894 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6LB72|C6LB72_9FIRM ABC transporter substrate-binding protein family 5 OS=Marvinbryantia formatexigens DSM 14469 OX=478749 GN=BRYFOR_05867 PE=4 SV=1
MM1 pKa = 7.76RR2 pKa = 11.84KK3 pKa = 9.94SSMKK7 pKa = 8.89ITGAVLCAAMCMSMTGGVSAEE28 pKa = 3.96EE29 pKa = 4.38TIEE32 pKa = 3.89VAYY35 pKa = 11.05NLTTEE40 pKa = 4.25DD41 pKa = 3.58FAVFEE46 pKa = 4.57QITKK50 pKa = 10.79DD51 pKa = 3.43FTEE54 pKa = 4.08EE55 pKa = 3.51TGIGVTIYY63 pKa = 10.99NGGDD67 pKa = 3.5DD68 pKa = 4.51YY69 pKa = 11.76EE70 pKa = 4.83SAMKK74 pKa = 10.03TRR76 pKa = 11.84MSSGDD81 pKa = 4.21LPDD84 pKa = 2.76MWTTHH89 pKa = 4.54GWSLIRR95 pKa = 11.84YY96 pKa = 8.23SEE98 pKa = 3.99YY99 pKa = 11.04MMDD102 pKa = 5.12LSDD105 pKa = 4.77QEE107 pKa = 4.35WVSNIDD113 pKa = 3.38EE114 pKa = 4.33GLKK117 pKa = 10.66NVITNDD123 pKa = 3.51DD124 pKa = 4.0GEE126 pKa = 4.92LFILPITQAVTAIVYY141 pKa = 10.39NKK143 pKa = 10.46DD144 pKa = 3.37VLDD147 pKa = 3.99EE148 pKa = 4.92AGVDD152 pKa = 3.58ASAIRR157 pKa = 11.84TWDD160 pKa = 3.63DD161 pKa = 3.11FDD163 pKa = 5.26AALQAVADD171 pKa = 4.06NTEE174 pKa = 3.95ATPLEE179 pKa = 4.17ICMGDD184 pKa = 3.54GYY186 pKa = 11.16DD187 pKa = 3.84SYY189 pKa = 11.55SLEE192 pKa = 5.15DD193 pKa = 3.41IWPTLYY199 pKa = 10.67TNSDD203 pKa = 3.48LADD206 pKa = 3.69NKK208 pKa = 11.24AEE210 pKa = 4.09TLQDD214 pKa = 3.52GTFDD218 pKa = 3.28WDD220 pKa = 3.33ADD222 pKa = 3.6GRR224 pKa = 11.84EE225 pKa = 4.35GFQMLADD232 pKa = 4.73WYY234 pKa = 9.15TKK236 pKa = 10.52GYY238 pKa = 9.3MNEE241 pKa = 4.61DD242 pKa = 3.64YY243 pKa = 11.52VSGKK247 pKa = 10.23KK248 pKa = 10.07DD249 pKa = 4.26DD250 pKa = 3.6ICKK253 pKa = 10.61AMANNEE259 pKa = 4.24GAFTLYY265 pKa = 8.39STEE268 pKa = 4.38MIPSVLAYY276 pKa = 10.69NEE278 pKa = 3.95NANLGILPVPSKK290 pKa = 10.46EE291 pKa = 4.12ADD293 pKa = 3.23GASYY297 pKa = 10.6FGTGEE302 pKa = 4.37GNFSCFGIWKK312 pKa = 8.87DD313 pKa = 3.8TEE315 pKa = 4.79HH316 pKa = 7.37EE317 pKa = 4.22DD318 pKa = 3.57ACKK321 pKa = 10.41QFLSYY326 pKa = 10.66LARR329 pKa = 11.84PEE331 pKa = 4.02VAVKK335 pKa = 10.26IVQIDD340 pKa = 3.52GGIPALTNIEE350 pKa = 4.45LDD352 pKa = 3.71EE353 pKa = 4.51TDD355 pKa = 3.13QAAYY359 pKa = 5.66TTNAFRR365 pKa = 11.84EE366 pKa = 4.32AQEE369 pKa = 4.14QFDD372 pKa = 4.18GDD374 pKa = 4.66LIYY377 pKa = 11.12DD378 pKa = 3.52NFFDD382 pKa = 5.47RR383 pKa = 11.84EE384 pKa = 4.15YY385 pKa = 10.24LPSGMWSVMSDD396 pKa = 3.05AVQVLLSDD404 pKa = 4.7GDD406 pKa = 4.19PEE408 pKa = 6.05ADD410 pKa = 3.11MDD412 pKa = 4.23EE413 pKa = 4.45AVMMVADD420 pKa = 4.82NYY422 pKa = 10.93NDD424 pKa = 3.81LMGNN428 pKa = 3.81
MM1 pKa = 7.76RR2 pKa = 11.84KK3 pKa = 9.94SSMKK7 pKa = 8.89ITGAVLCAAMCMSMTGGVSAEE28 pKa = 3.96EE29 pKa = 4.38TIEE32 pKa = 3.89VAYY35 pKa = 11.05NLTTEE40 pKa = 4.25DD41 pKa = 3.58FAVFEE46 pKa = 4.57QITKK50 pKa = 10.79DD51 pKa = 3.43FTEE54 pKa = 4.08EE55 pKa = 3.51TGIGVTIYY63 pKa = 10.99NGGDD67 pKa = 3.5DD68 pKa = 4.51YY69 pKa = 11.76EE70 pKa = 4.83SAMKK74 pKa = 10.03TRR76 pKa = 11.84MSSGDD81 pKa = 4.21LPDD84 pKa = 2.76MWTTHH89 pKa = 4.54GWSLIRR95 pKa = 11.84YY96 pKa = 8.23SEE98 pKa = 3.99YY99 pKa = 11.04MMDD102 pKa = 5.12LSDD105 pKa = 4.77QEE107 pKa = 4.35WVSNIDD113 pKa = 3.38EE114 pKa = 4.33GLKK117 pKa = 10.66NVITNDD123 pKa = 3.51DD124 pKa = 4.0GEE126 pKa = 4.92LFILPITQAVTAIVYY141 pKa = 10.39NKK143 pKa = 10.46DD144 pKa = 3.37VLDD147 pKa = 3.99EE148 pKa = 4.92AGVDD152 pKa = 3.58ASAIRR157 pKa = 11.84TWDD160 pKa = 3.63DD161 pKa = 3.11FDD163 pKa = 5.26AALQAVADD171 pKa = 4.06NTEE174 pKa = 3.95ATPLEE179 pKa = 4.17ICMGDD184 pKa = 3.54GYY186 pKa = 11.16DD187 pKa = 3.84SYY189 pKa = 11.55SLEE192 pKa = 5.15DD193 pKa = 3.41IWPTLYY199 pKa = 10.67TNSDD203 pKa = 3.48LADD206 pKa = 3.69NKK208 pKa = 11.24AEE210 pKa = 4.09TLQDD214 pKa = 3.52GTFDD218 pKa = 3.28WDD220 pKa = 3.33ADD222 pKa = 3.6GRR224 pKa = 11.84EE225 pKa = 4.35GFQMLADD232 pKa = 4.73WYY234 pKa = 9.15TKK236 pKa = 10.52GYY238 pKa = 9.3MNEE241 pKa = 4.61DD242 pKa = 3.64YY243 pKa = 11.52VSGKK247 pKa = 10.23KK248 pKa = 10.07DD249 pKa = 4.26DD250 pKa = 3.6ICKK253 pKa = 10.61AMANNEE259 pKa = 4.24GAFTLYY265 pKa = 8.39STEE268 pKa = 4.38MIPSVLAYY276 pKa = 10.69NEE278 pKa = 3.95NANLGILPVPSKK290 pKa = 10.46EE291 pKa = 4.12ADD293 pKa = 3.23GASYY297 pKa = 10.6FGTGEE302 pKa = 4.37GNFSCFGIWKK312 pKa = 8.87DD313 pKa = 3.8TEE315 pKa = 4.79HH316 pKa = 7.37EE317 pKa = 4.22DD318 pKa = 3.57ACKK321 pKa = 10.41QFLSYY326 pKa = 10.66LARR329 pKa = 11.84PEE331 pKa = 4.02VAVKK335 pKa = 10.26IVQIDD340 pKa = 3.52GGIPALTNIEE350 pKa = 4.45LDD352 pKa = 3.71EE353 pKa = 4.51TDD355 pKa = 3.13QAAYY359 pKa = 5.66TTNAFRR365 pKa = 11.84EE366 pKa = 4.32AQEE369 pKa = 4.14QFDD372 pKa = 4.18GDD374 pKa = 4.66LIYY377 pKa = 11.12DD378 pKa = 3.52NFFDD382 pKa = 5.47RR383 pKa = 11.84EE384 pKa = 4.15YY385 pKa = 10.24LPSGMWSVMSDD396 pKa = 3.05AVQVLLSDD404 pKa = 4.7GDD406 pKa = 4.19PEE408 pKa = 6.05ADD410 pKa = 3.11MDD412 pKa = 4.23EE413 pKa = 4.45AVMMVADD420 pKa = 4.82NYY422 pKa = 10.93NDD424 pKa = 3.81LMGNN428 pKa = 3.81
Molecular weight: 47.39 kDa
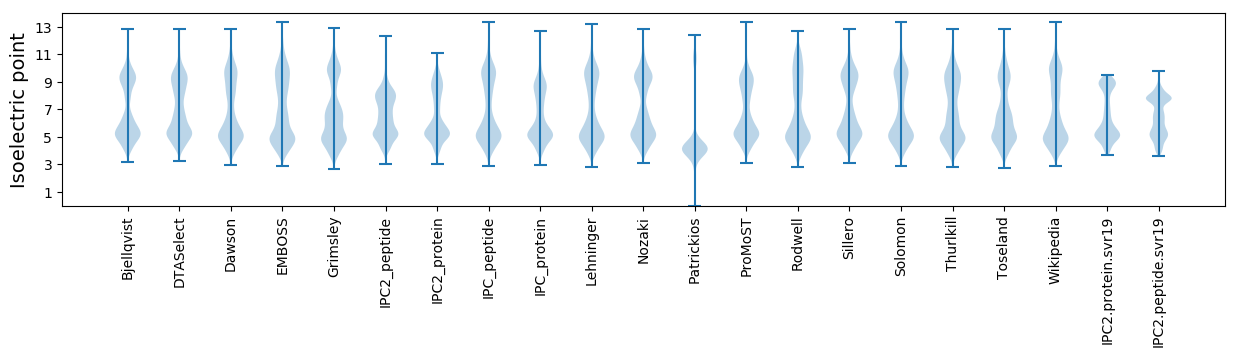
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6LKL5|C6LKL5_9FIRM Uncharacterized protein OS=Marvinbryantia formatexigens DSM 14469 OX=478749 GN=BRYFOR_09203 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.29GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.5TAGGRR28 pKa = 11.84KK29 pKa = 8.77VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.75GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.29GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.5TAGGRR28 pKa = 11.84KK29 pKa = 8.77VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.75GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 5.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1389242 |
36 |
2109 |
283.9 |
31.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.243 ± 0.047 | 1.621 ± 0.017 |
5.329 ± 0.031 | 7.72 ± 0.044 |
4.102 ± 0.028 | 7.316 ± 0.037 |
1.699 ± 0.017 | 6.783 ± 0.034 |
6.059 ± 0.031 | 8.995 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.145 ± 0.019 | 3.951 ± 0.023 |
3.389 ± 0.022 | 3.429 ± 0.022 |
5.06 ± 0.038 | 5.682 ± 0.029 |
5.544 ± 0.038 | 6.713 ± 0.027 |
1.046 ± 0.012 | 4.173 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
