
Absidia glauca (Pin mould)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Mucoromycota; Mucoromycotina; Mucoromycetes; Mucorales; Cunninghamellaceae; Absidia
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
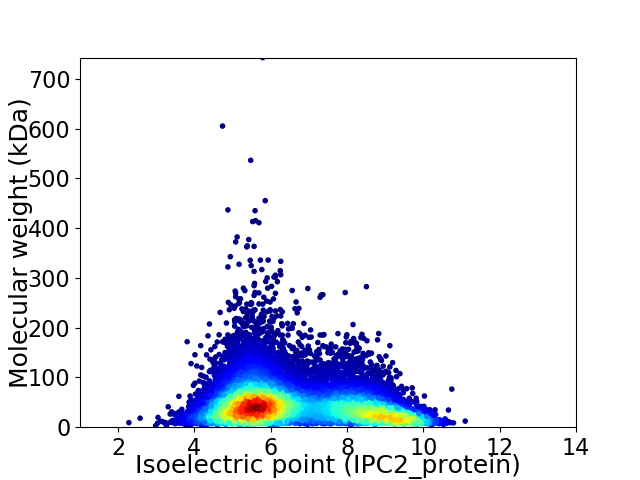
Virtual 2D-PAGE plot for 14825 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A163JZ98|A0A163JZ98_ABSGL Uncharacterized protein OS=Absidia glauca OX=4829 GN=ABSGL_11451.1 scaffold 12295 PE=4 SV=1
MM1 pKa = 7.77PALTLDD7 pKa = 3.74RR8 pKa = 11.84LQAAVCFLKK17 pKa = 10.47MSLDD21 pKa = 3.54DD22 pKa = 3.91HH23 pKa = 6.4GVRR26 pKa = 11.84FAPPLSAQALEE37 pKa = 4.26KK38 pKa = 10.65FEE40 pKa = 5.1CEE42 pKa = 3.79HH43 pKa = 5.71TVRR46 pKa = 11.84LPEE49 pKa = 4.53DD50 pKa = 3.26YY51 pKa = 10.93RR52 pKa = 11.84FFLTLIGNGGQSGPCFEE69 pKa = 4.87GVLPLGVFPFTSNPEE84 pKa = 3.96SLLNNLPDD92 pKa = 4.39LFPFTEE98 pKa = 4.2PSILIGGGGGQQQDD112 pKa = 3.91SDD114 pKa = 5.39DD115 pKa = 5.55DD116 pKa = 4.09DD117 pKa = 4.69TDD119 pKa = 4.35LCDD122 pKa = 3.68TTSDD126 pKa = 4.15DD127 pKa = 4.56NGASTDD133 pKa = 3.78DD134 pKa = 4.0SLDD137 pKa = 4.22GILGNTYY144 pKa = 10.56SSSDD148 pKa = 3.27NDD150 pKa = 4.61DD151 pKa = 4.74RR152 pKa = 11.84EE153 pKa = 4.42DD154 pKa = 5.56DD155 pKa = 3.85YY156 pKa = 12.1DD157 pKa = 6.39DD158 pKa = 5.94YY159 pKa = 12.09DD160 pKa = 6.76DD161 pKa = 6.84DD162 pKa = 7.48DD163 pKa = 7.23DD164 pKa = 7.7DD165 pKa = 5.4DD166 pKa = 4.82DD167 pKa = 6.75KK168 pKa = 12.26YY169 pKa = 8.86MTSYY173 pKa = 11.45SLDD176 pKa = 3.41EE177 pKa = 4.07EE178 pKa = 5.42LYY180 pKa = 10.16PNLQVVEE187 pKa = 4.21QQQQQQRR194 pKa = 11.84DD195 pKa = 3.63GSIKK199 pKa = 10.83HH200 pKa = 6.4NDD202 pKa = 2.92TMTVGGYY209 pKa = 10.61LILGTLAQDD218 pKa = 3.57RR219 pKa = 11.84QYY221 pKa = 11.45FWILICGGACRR232 pKa = 11.84GEE234 pKa = 3.7VWIMNKK240 pKa = 10.0HH241 pKa = 5.25GFYY244 pKa = 10.54FPTSRR249 pKa = 11.84RR250 pKa = 11.84LKK252 pKa = 8.97FYY254 pKa = 10.57EE255 pKa = 4.2WVEE258 pKa = 3.91DD259 pKa = 3.58WLLGGDD265 pKa = 4.99KK266 pKa = 10.55INASLWACGLYY277 pKa = 10.52DD278 pKa = 3.94PQFSRR283 pKa = 11.84CFQSGIDD290 pKa = 3.7NATSDD295 pKa = 3.62SGSDD299 pKa = 3.4QEE301 pKa = 4.29EE302 pKa = 4.96DD303 pKa = 3.72YY304 pKa = 11.77GNSSDD309 pKa = 3.93NDD311 pKa = 4.04SNSGYY316 pKa = 10.82SYY318 pKa = 11.39DD319 pKa = 5.36DD320 pKa = 3.51EE321 pKa = 4.6LL322 pKa = 6.09
MM1 pKa = 7.77PALTLDD7 pKa = 3.74RR8 pKa = 11.84LQAAVCFLKK17 pKa = 10.47MSLDD21 pKa = 3.54DD22 pKa = 3.91HH23 pKa = 6.4GVRR26 pKa = 11.84FAPPLSAQALEE37 pKa = 4.26KK38 pKa = 10.65FEE40 pKa = 5.1CEE42 pKa = 3.79HH43 pKa = 5.71TVRR46 pKa = 11.84LPEE49 pKa = 4.53DD50 pKa = 3.26YY51 pKa = 10.93RR52 pKa = 11.84FFLTLIGNGGQSGPCFEE69 pKa = 4.87GVLPLGVFPFTSNPEE84 pKa = 3.96SLLNNLPDD92 pKa = 4.39LFPFTEE98 pKa = 4.2PSILIGGGGGQQQDD112 pKa = 3.91SDD114 pKa = 5.39DD115 pKa = 5.55DD116 pKa = 4.09DD117 pKa = 4.69TDD119 pKa = 4.35LCDD122 pKa = 3.68TTSDD126 pKa = 4.15DD127 pKa = 4.56NGASTDD133 pKa = 3.78DD134 pKa = 4.0SLDD137 pKa = 4.22GILGNTYY144 pKa = 10.56SSSDD148 pKa = 3.27NDD150 pKa = 4.61DD151 pKa = 4.74RR152 pKa = 11.84EE153 pKa = 4.42DD154 pKa = 5.56DD155 pKa = 3.85YY156 pKa = 12.1DD157 pKa = 6.39DD158 pKa = 5.94YY159 pKa = 12.09DD160 pKa = 6.76DD161 pKa = 6.84DD162 pKa = 7.48DD163 pKa = 7.23DD164 pKa = 7.7DD165 pKa = 5.4DD166 pKa = 4.82DD167 pKa = 6.75KK168 pKa = 12.26YY169 pKa = 8.86MTSYY173 pKa = 11.45SLDD176 pKa = 3.41EE177 pKa = 4.07EE178 pKa = 5.42LYY180 pKa = 10.16PNLQVVEE187 pKa = 4.21QQQQQQRR194 pKa = 11.84DD195 pKa = 3.63GSIKK199 pKa = 10.83HH200 pKa = 6.4NDD202 pKa = 2.92TMTVGGYY209 pKa = 10.61LILGTLAQDD218 pKa = 3.57RR219 pKa = 11.84QYY221 pKa = 11.45FWILICGGACRR232 pKa = 11.84GEE234 pKa = 3.7VWIMNKK240 pKa = 10.0HH241 pKa = 5.25GFYY244 pKa = 10.54FPTSRR249 pKa = 11.84RR250 pKa = 11.84LKK252 pKa = 8.97FYY254 pKa = 10.57EE255 pKa = 4.2WVEE258 pKa = 3.91DD259 pKa = 3.58WLLGGDD265 pKa = 4.99KK266 pKa = 10.55INASLWACGLYY277 pKa = 10.52DD278 pKa = 3.94PQFSRR283 pKa = 11.84CFQSGIDD290 pKa = 3.7NATSDD295 pKa = 3.62SGSDD299 pKa = 3.4QEE301 pKa = 4.29EE302 pKa = 4.96DD303 pKa = 3.72YY304 pKa = 11.77GNSSDD309 pKa = 3.93NDD311 pKa = 4.04SNSGYY316 pKa = 10.82SYY318 pKa = 11.39DD319 pKa = 5.36DD320 pKa = 3.51EE321 pKa = 4.6LL322 pKa = 6.09
Molecular weight: 35.97 kDa
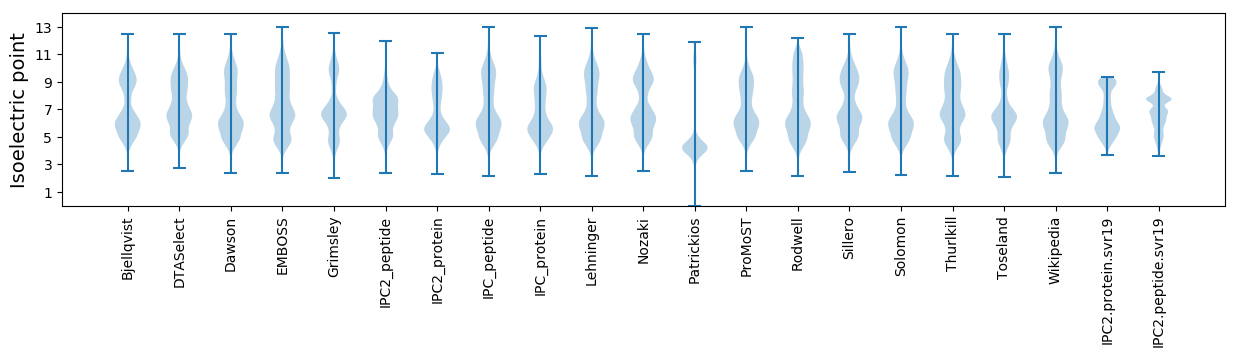
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168RTF4|A0A168RTF4_ABSGL DNA polymerase epsilon subunit OS=Absidia glauca OX=4829 GN=ABSGL_13010.1 scaffold 13554 PE=3 SV=1
MM1 pKa = 7.95DD2 pKa = 4.8FSSHH6 pKa = 6.5RR7 pKa = 11.84EE8 pKa = 4.16SIGKK12 pKa = 7.4TFSSPGIRR20 pKa = 11.84SNKK23 pKa = 9.18KK24 pKa = 8.4IHH26 pKa = 6.43INCGSSARR34 pKa = 11.84MAGNMCANVDD44 pKa = 3.62QIRR47 pKa = 11.84RR48 pKa = 11.84QGRR51 pKa = 11.84RR52 pKa = 11.84DD53 pKa = 3.1NTTINGAYY61 pKa = 7.77LTNLPRR67 pKa = 11.84EE68 pKa = 4.67LVRR71 pKa = 11.84SMAGSLTNGRR81 pKa = 11.84FFYY84 pKa = 10.72LARR87 pKa = 11.84AALNPCGIARR97 pKa = 11.84EE98 pKa = 4.21EE99 pKa = 4.05
MM1 pKa = 7.95DD2 pKa = 4.8FSSHH6 pKa = 6.5RR7 pKa = 11.84EE8 pKa = 4.16SIGKK12 pKa = 7.4TFSSPGIRR20 pKa = 11.84SNKK23 pKa = 9.18KK24 pKa = 8.4IHH26 pKa = 6.43INCGSSARR34 pKa = 11.84MAGNMCANVDD44 pKa = 3.62QIRR47 pKa = 11.84RR48 pKa = 11.84QGRR51 pKa = 11.84RR52 pKa = 11.84DD53 pKa = 3.1NTTINGAYY61 pKa = 7.77LTNLPRR67 pKa = 11.84EE68 pKa = 4.67LVRR71 pKa = 11.84SMAGSLTNGRR81 pKa = 11.84FFYY84 pKa = 10.72LARR87 pKa = 11.84AALNPCGIARR97 pKa = 11.84EE98 pKa = 4.21EE99 pKa = 4.05
Molecular weight: 10.94 kDa
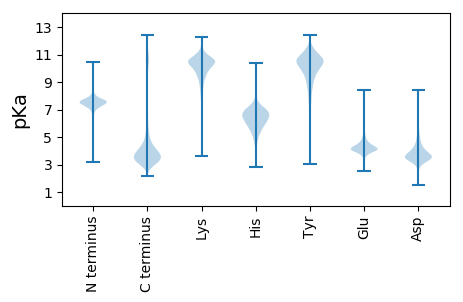
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6549974 |
19 |
6879 |
441.8 |
49.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.916 ± 0.018 | 1.377 ± 0.008 |
6.274 ± 0.016 | 5.082 ± 0.017 |
3.652 ± 0.012 | 5.412 ± 0.018 |
3.053 ± 0.012 | 5.141 ± 0.013 |
5.315 ± 0.017 | 9.318 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.538 ± 0.007 | 4.323 ± 0.011 |
5.635 ± 0.023 | 5.151 ± 0.021 |
5.163 ± 0.014 | 8.663 ± 0.027 |
6.779 ± 0.016 | 5.751 ± 0.017 |
1.213 ± 0.006 | 3.182 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
