
Wuhan Louse Fly Virus 9
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sigmavirus; Hippoboscid sigmavirus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
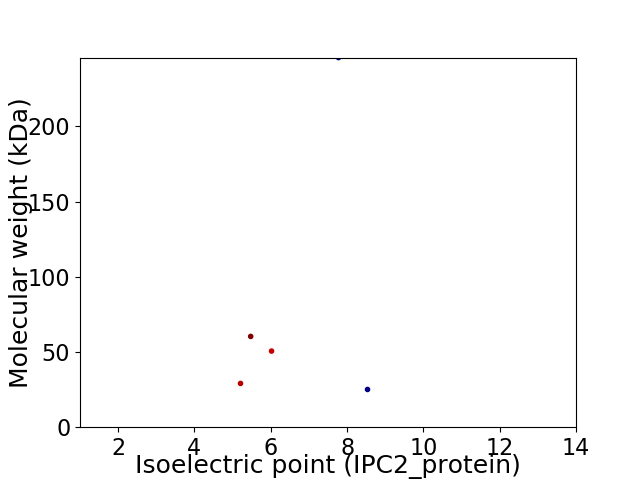
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KKB5|A0A0B5KKB5_9VIRU Matrix protein OS=Wuhan Louse Fly Virus 9 OX=1608123 GN=M PE=4 SV=1
MM1 pKa = 7.84EE2 pKa = 5.83SPNQDD7 pKa = 2.5SDD9 pKa = 4.33LEE11 pKa = 4.14EE12 pKa = 4.37MQDD15 pKa = 3.86KK16 pKa = 10.77LNKK19 pKa = 10.48LEE21 pKa = 4.7DD22 pKa = 3.81LDD24 pKa = 3.94VFKK27 pKa = 10.7EE28 pKa = 4.24IEE30 pKa = 4.15AQTQTNAADD39 pKa = 3.32WSIDD43 pKa = 3.22GKK45 pKa = 11.26KK46 pKa = 10.07LVPTDD51 pKa = 4.23FDD53 pKa = 4.79SDD55 pKa = 3.82SSDD58 pKa = 4.02YY59 pKa = 11.46EE60 pKa = 4.14DD61 pKa = 6.27DD62 pKa = 3.79EE63 pKa = 4.38ALKK66 pKa = 11.05LGTTILAKK74 pKa = 10.23PKK76 pKa = 10.44LIVPPPQDD84 pKa = 3.26EE85 pKa = 4.28RR86 pKa = 11.84QGPSGISFTPLQQTYY101 pKa = 9.1TDD103 pKa = 3.8YY104 pKa = 11.06QQSVKK109 pKa = 9.43QKK111 pKa = 10.23RR112 pKa = 11.84IRR114 pKa = 11.84TMTTGANYY122 pKa = 10.42NIVVNDD128 pKa = 3.74PRR130 pKa = 11.84FLVCPSDD137 pKa = 3.49LCNEE141 pKa = 4.23LGFLFYY147 pKa = 10.86AVATRR152 pKa = 11.84AIRR155 pKa = 11.84PEE157 pKa = 3.97QVCLYY162 pKa = 9.12FNSGGTQTMHH172 pKa = 6.34QGAGLNYY179 pKa = 10.09NIFISGLQQQLPPPQFPPPPPPPIPRR205 pKa = 11.84CSPILSNLPSLDD217 pKa = 3.4KK218 pKa = 10.87LIRR221 pKa = 11.84AADD224 pKa = 4.09PFPSHH229 pKa = 7.17PFVKK233 pKa = 10.31KK234 pKa = 9.72LQEE237 pKa = 4.55GIKK240 pKa = 10.3LKK242 pKa = 10.6HH243 pKa = 6.32LYY245 pKa = 9.98LSKK248 pKa = 10.03KK249 pKa = 8.37TNNDD253 pKa = 2.63LHH255 pKa = 7.49KK256 pKa = 10.39PAGFF260 pKa = 3.66
MM1 pKa = 7.84EE2 pKa = 5.83SPNQDD7 pKa = 2.5SDD9 pKa = 4.33LEE11 pKa = 4.14EE12 pKa = 4.37MQDD15 pKa = 3.86KK16 pKa = 10.77LNKK19 pKa = 10.48LEE21 pKa = 4.7DD22 pKa = 3.81LDD24 pKa = 3.94VFKK27 pKa = 10.7EE28 pKa = 4.24IEE30 pKa = 4.15AQTQTNAADD39 pKa = 3.32WSIDD43 pKa = 3.22GKK45 pKa = 11.26KK46 pKa = 10.07LVPTDD51 pKa = 4.23FDD53 pKa = 4.79SDD55 pKa = 3.82SSDD58 pKa = 4.02YY59 pKa = 11.46EE60 pKa = 4.14DD61 pKa = 6.27DD62 pKa = 3.79EE63 pKa = 4.38ALKK66 pKa = 11.05LGTTILAKK74 pKa = 10.23PKK76 pKa = 10.44LIVPPPQDD84 pKa = 3.26EE85 pKa = 4.28RR86 pKa = 11.84QGPSGISFTPLQQTYY101 pKa = 9.1TDD103 pKa = 3.8YY104 pKa = 11.06QQSVKK109 pKa = 9.43QKK111 pKa = 10.23RR112 pKa = 11.84IRR114 pKa = 11.84TMTTGANYY122 pKa = 10.42NIVVNDD128 pKa = 3.74PRR130 pKa = 11.84FLVCPSDD137 pKa = 3.49LCNEE141 pKa = 4.23LGFLFYY147 pKa = 10.86AVATRR152 pKa = 11.84AIRR155 pKa = 11.84PEE157 pKa = 3.97QVCLYY162 pKa = 9.12FNSGGTQTMHH172 pKa = 6.34QGAGLNYY179 pKa = 10.09NIFISGLQQQLPPPQFPPPPPPPIPRR205 pKa = 11.84CSPILSNLPSLDD217 pKa = 3.4KK218 pKa = 10.87LIRR221 pKa = 11.84AADD224 pKa = 4.09PFPSHH229 pKa = 7.17PFVKK233 pKa = 10.31KK234 pKa = 9.72LQEE237 pKa = 4.55GIKK240 pKa = 10.3LKK242 pKa = 10.6HH243 pKa = 6.32LYY245 pKa = 9.98LSKK248 pKa = 10.03KK249 pKa = 8.37TNNDD253 pKa = 2.63LHH255 pKa = 7.49KK256 pKa = 10.39PAGFF260 pKa = 3.66
Molecular weight: 29.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KRS6|A0A0B5KRS6_9VIRU Glycoprotein OS=Wuhan Louse Fly Virus 9 OX=1608123 GN=G PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84MKK4 pKa = 10.25KK5 pKa = 10.08VKK7 pKa = 10.55NLVSPRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 10.04AKK17 pKa = 10.39SDD19 pKa = 3.38EE20 pKa = 4.39SNQASAVITPTAPSPSSTTPFSVPTIQPNIVPSIITSRR58 pKa = 11.84WNTVGSLRR66 pKa = 11.84ISCEE70 pKa = 3.7RR71 pKa = 11.84EE72 pKa = 3.41ITDD75 pKa = 3.23WVTMEE80 pKa = 4.45RR81 pKa = 11.84LLEE84 pKa = 4.0YY85 pKa = 10.41MLDD88 pKa = 3.76FYY90 pKa = 11.6DD91 pKa = 4.01GNEE94 pKa = 3.82VFKK97 pKa = 10.65PVILMLYY104 pKa = 8.8WILGVHH110 pKa = 6.78LKK112 pKa = 10.29PVEE115 pKa = 4.42GPSNRR120 pKa = 11.84WCWGIDD126 pKa = 3.28FGEE129 pKa = 4.59PIEE132 pKa = 4.43VKK134 pKa = 10.53HH135 pKa = 6.04KK136 pKa = 10.5FPLIGSQSFDD146 pKa = 3.11YY147 pKa = 10.66RR148 pKa = 11.84KK149 pKa = 10.59NITSNYY155 pKa = 9.75RR156 pKa = 11.84GIKK159 pKa = 9.97FNIEE163 pKa = 3.28FWIQFTGTQRR173 pKa = 11.84RR174 pKa = 11.84STPVKK179 pKa = 10.26DD180 pKa = 3.63LLGVGSLIFPDD191 pKa = 3.22VRR193 pKa = 11.84RR194 pKa = 11.84FKK196 pKa = 10.84EE197 pKa = 3.84ILDD200 pKa = 3.77FSNINCEE207 pKa = 3.82LDD209 pKa = 3.66SNKK212 pKa = 10.24NLVLTQQ218 pKa = 3.44
MM1 pKa = 7.9RR2 pKa = 11.84MKK4 pKa = 10.25KK5 pKa = 10.08VKK7 pKa = 10.55NLVSPRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 10.04AKK17 pKa = 10.39SDD19 pKa = 3.38EE20 pKa = 4.39SNQASAVITPTAPSPSSTTPFSVPTIQPNIVPSIITSRR58 pKa = 11.84WNTVGSLRR66 pKa = 11.84ISCEE70 pKa = 3.7RR71 pKa = 11.84EE72 pKa = 3.41ITDD75 pKa = 3.23WVTMEE80 pKa = 4.45RR81 pKa = 11.84LLEE84 pKa = 4.0YY85 pKa = 10.41MLDD88 pKa = 3.76FYY90 pKa = 11.6DD91 pKa = 4.01GNEE94 pKa = 3.82VFKK97 pKa = 10.65PVILMLYY104 pKa = 8.8WILGVHH110 pKa = 6.78LKK112 pKa = 10.29PVEE115 pKa = 4.42GPSNRR120 pKa = 11.84WCWGIDD126 pKa = 3.28FGEE129 pKa = 4.59PIEE132 pKa = 4.43VKK134 pKa = 10.53HH135 pKa = 6.04KK136 pKa = 10.5FPLIGSQSFDD146 pKa = 3.11YY147 pKa = 10.66RR148 pKa = 11.84KK149 pKa = 10.59NITSNYY155 pKa = 9.75RR156 pKa = 11.84GIKK159 pKa = 9.97FNIEE163 pKa = 3.28FWIQFTGTQRR173 pKa = 11.84RR174 pKa = 11.84STPVKK179 pKa = 10.26DD180 pKa = 3.63LLGVGSLIFPDD191 pKa = 3.22VRR193 pKa = 11.84RR194 pKa = 11.84FKK196 pKa = 10.84EE197 pKa = 3.84ILDD200 pKa = 3.77FSNINCEE207 pKa = 3.82LDD209 pKa = 3.66SNKK212 pKa = 10.24NLVLTQQ218 pKa = 3.44
Molecular weight: 25.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3604 |
218 |
2145 |
720.8 |
82.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.635 ± 0.388 | 1.609 ± 0.145 |
5.161 ± 0.427 | 5.522 ± 0.54 |
4.412 ± 0.261 | 5.272 ± 0.28 |
2.58 ± 0.331 | 7.714 ± 0.274 |
6.243 ± 0.303 | 11.043 ± 0.713 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.831 ± 0.348 | 5.716 ± 0.26 |
5.3 ± 0.928 | 3.857 ± 0.427 |
4.55 ± 0.558 | 8.546 ± 0.294 |
5.966 ± 0.624 | 5.66 ± 0.659 |
1.693 ± 0.173 | 3.69 ± 0.247 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
