
Sphingopyxis sp. LPB0140
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingopyxis; unclassified Sphingopyxis
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
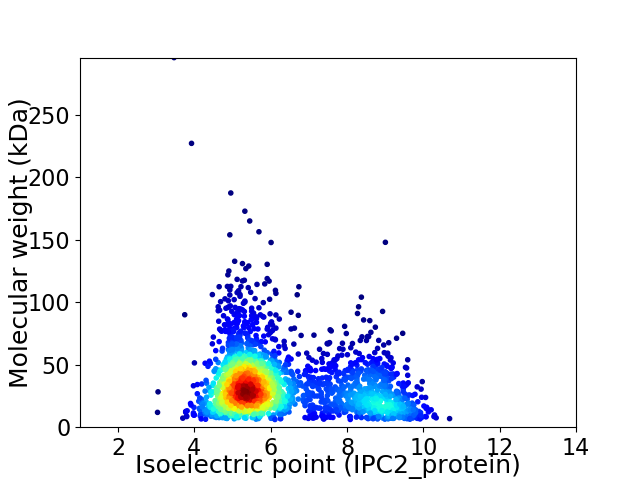
Virtual 2D-PAGE plot for 2350 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3J9A9|A0A1L3J9A9_9SPHN Aldo/keto reductase OS=Sphingopyxis sp. LPB0140 OX=1913578 GN=LPB140_01225 PE=4 SV=1
MM1 pKa = 7.53RR2 pKa = 11.84HH3 pKa = 6.14LAFDD7 pKa = 3.56NQKK10 pKa = 8.29TVISSSEE17 pKa = 3.83QARR20 pKa = 11.84KK21 pKa = 9.25LLKK24 pKa = 9.34RR25 pKa = 11.84AKK27 pKa = 8.39WLAGASILAASSTAMAAPSLVVNNVGTPTVVGTGQGKK64 pKa = 9.02RR65 pKa = 11.84AIWTNAGTVGSDD77 pKa = 3.32TIDD80 pKa = 3.31LVGVITTATLNHH92 pKa = 6.18TWSTTANRR100 pKa = 11.84PSITSVGPDD109 pKa = 3.78DD110 pKa = 5.92IFIEE114 pKa = 3.97WRR116 pKa = 11.84LYY118 pKa = 10.42RR119 pKa = 11.84AGTYY123 pKa = 10.58NITTNSGGVPVVADD137 pKa = 2.84VHH139 pKa = 6.12VQFNDD144 pKa = 2.69VDD146 pKa = 3.86GPNNEE151 pKa = 4.33RR152 pKa = 11.84IYY154 pKa = 11.23LPVCQGDD161 pKa = 3.83ISWVRR166 pKa = 11.84IDD168 pKa = 3.49ATATTGRR175 pKa = 11.84AFGAVAGQAEE185 pKa = 4.88TFSLIGDD192 pKa = 3.56QNYY195 pKa = 10.22NNQPVSGLEE204 pKa = 3.95ALWKK208 pKa = 10.51DD209 pKa = 2.99RR210 pKa = 11.84STFTMGRR217 pKa = 11.84TANSGFLIRR226 pKa = 11.84FDD228 pKa = 3.77NPTYY232 pKa = 10.88SAFDD236 pKa = 3.57TLDD239 pKa = 3.52FEE241 pKa = 5.23CADD244 pKa = 4.22FKK246 pKa = 11.52PPVTVNDD253 pKa = 3.84SKK255 pKa = 11.45EE256 pKa = 4.25GVPGTSTIVDD266 pKa = 3.27ILDD269 pKa = 3.9NDD271 pKa = 4.16SSATLNNNPTNNNSLKK287 pKa = 10.55ASEE290 pKa = 4.23FARR293 pKa = 11.84ASVNLVPPPSATGIITDD310 pKa = 3.69SFGDD314 pKa = 3.63VIGFTVPGEE323 pKa = 4.51GVWSYY328 pKa = 12.09SDD330 pKa = 3.21TTGQLTFTPDD340 pKa = 3.3PSFVGYY346 pKa = 10.81ASTVNYY352 pKa = 9.32TVDD355 pKa = 3.26NALGIQSNQATVTVWYY371 pKa = 9.12PGIGVTKK378 pKa = 10.66ASTFNDD384 pKa = 3.76LNGDD388 pKa = 3.61GYY390 pKa = 10.93GQVGEE395 pKa = 4.53TINYY399 pKa = 7.86IYY401 pKa = 9.95QVKK404 pKa = 10.36SYY406 pKa = 10.57GAEE409 pKa = 3.93PLRR412 pKa = 11.84NPSLTEE418 pKa = 3.81TTFTGAGTPPTPTYY432 pKa = 11.2VSGDD436 pKa = 3.87TNNDD440 pKa = 2.99GYY442 pKa = 10.71IGLTEE447 pKa = 3.83TWFYY451 pKa = 10.73SAVYY455 pKa = 10.3SLVAGDD461 pKa = 4.36LSGTGVNNSATASGNTAAGTVVNDD485 pKa = 3.86VSDD488 pKa = 3.85SANPADD494 pKa = 4.3GNNNTKK500 pKa = 10.45SGPGPGNNDD509 pKa = 2.87ATVTAVPRR517 pKa = 11.84APIAASNDD525 pKa = 3.88TQSGTITNDD534 pKa = 2.97GQANAFNVLSNDD546 pKa = 3.61SLKK549 pKa = 11.01GAAPTPSNVAITVTTPASHH568 pKa = 7.13AGVTLNTSTGQVSVAPGTPAGNYY591 pKa = 6.86TINYY595 pKa = 6.67QICEE599 pKa = 3.9IGNPTNCANAVATVNVSLRR618 pKa = 11.84PINADD623 pKa = 3.43DD624 pKa = 4.91DD625 pKa = 4.5SGPSVNSTNGSASLYY640 pKa = 10.37NVLSNDD646 pKa = 3.5TFNGGAATTPNVTINVTSPATPAFAGASVPSLNISTGVVSVPAGTPAGTYY696 pKa = 9.9NIGYY700 pKa = 7.32EE701 pKa = 4.17ICDD704 pKa = 3.39SAVPTNCEE712 pKa = 3.33AAIATIIVTPTPIAASNDD730 pKa = 3.13SGPAVNGAAGNTNAFNAFTNDD751 pKa = 3.37SFNGNPVNLSLVTATVTSPATPLSAGAPVPSLNTATGVVSVPAGTPAGSYY801 pKa = 8.84TIGYY805 pKa = 7.73RR806 pKa = 11.84ICEE809 pKa = 4.05NANPTNCANASITVQVAASPIAANNDD835 pKa = 2.96STPAVNSANGGSNLVNALGNDD856 pKa = 4.04SLNGAAPTASNVNITVTTPASNAGVTMNTATGQVSVAPGTPAGNYY901 pKa = 9.64SIGYY905 pKa = 7.13QICEE909 pKa = 4.1KK910 pKa = 10.7LNPSNCAIANVTIPVNVTPIAASADD935 pKa = 3.69TPPAQNGRR943 pKa = 11.84TGANDD948 pKa = 2.91IVNAFTNDD956 pKa = 3.54SLNGSPVNVADD967 pKa = 3.91IEE969 pKa = 4.32ATITAPATPLTAGAPVPVMDD989 pKa = 5.18PATGLVDD996 pKa = 4.08VPAGTPAGTYY1006 pKa = 8.56TIAYY1010 pKa = 7.19EE1011 pKa = 4.12ICEE1014 pKa = 4.19KK1015 pKa = 10.45TNPTNCANSSVTVIVSAAPITALNDD1040 pKa = 3.25NAGTVTTANGGANLINALGNDD1061 pKa = 4.16TLNGSPVALADD1072 pKa = 3.64VNLTVTAPASNAGVTLNTATGQVSVAPGTPAGNYY1106 pKa = 9.64SIGYY1110 pKa = 7.13QICEE1114 pKa = 4.17KK1115 pKa = 10.75LNPTNCQSATIAVVVDD1131 pKa = 4.03VTPLNASNDD1140 pKa = 3.23SPAPVNGANGGNDD1153 pKa = 3.64IINAFANDD1161 pKa = 4.2TLNGAPVNVADD1172 pKa = 4.75ISATITAPATPLTAGAPVPVMDD1194 pKa = 5.18PATGLVDD1201 pKa = 4.08VPAGTPAGTYY1211 pKa = 8.56TIAYY1215 pKa = 7.18EE1216 pKa = 3.77ICEE1219 pKa = 4.08NSNPTNCKK1227 pKa = 7.55TATVQVVVTAAPITADD1243 pKa = 3.68ADD1245 pKa = 3.64SPAPVNGATGGNDD1258 pKa = 3.53IINAFANDD1266 pKa = 4.17SLNGAPVNVADD1277 pKa = 4.75IEE1279 pKa = 4.43ATVTSPAMPLTAGAPVPVMDD1299 pKa = 5.18PATGLVDD1306 pKa = 4.08VPAGTPAGTYY1316 pKa = 8.56TIAYY1320 pKa = 7.25EE1321 pKa = 4.01ICEE1324 pKa = 4.09KK1325 pKa = 10.94LNPNNCASSTVNVVVTAAPITADD1348 pKa = 3.68ADD1350 pKa = 3.64SPAPVNGATGGNDD1363 pKa = 3.53IINAFANDD1371 pKa = 4.17SLNGAPVNVADD1382 pKa = 4.75IEE1384 pKa = 4.43ATVTSPATPLTAGAPVPVMDD1404 pKa = 5.18PATGLVDD1411 pKa = 3.83VPVGTPAGTYY1421 pKa = 8.32TIAYY1425 pKa = 7.25EE1426 pKa = 4.01ICEE1429 pKa = 4.09KK1430 pKa = 10.94LNPNNCATNNVTVVVTAAPIAASNDD1455 pKa = 3.02AVGPVPGISGGSNLINALNNDD1476 pKa = 4.15MLNGASVAIADD1487 pKa = 3.74INVTVTSPASHH1498 pKa = 6.82AGVTLDD1504 pKa = 4.06PATGMVSVAPNTPAGTYY1521 pKa = 9.0TIGYY1525 pKa = 7.18QICEE1529 pKa = 4.21KK1530 pKa = 10.84LNPTNCANAIISVTVDD1546 pKa = 3.73AAPIMADD1553 pKa = 3.01VDD1555 pKa = 4.0SPAPVNGANGGNDD1568 pKa = 3.64IINAFANDD1576 pKa = 4.17SLNGAPVNVADD1587 pKa = 4.46IDD1589 pKa = 3.98ATITTPATPATPGAPVPVMDD1609 pKa = 5.18PATGLVDD1616 pKa = 3.54VSVGTPAGTYY1626 pKa = 8.32TIAYY1630 pKa = 7.51EE1631 pKa = 4.19ICQKK1635 pKa = 10.92LNPTNCASSNVTIVVEE1651 pKa = 4.14APEE1654 pKa = 3.83ILAANDD1660 pKa = 2.82VPAPVRR1666 pKa = 11.84SGVGNPNAINAFANDD1681 pKa = 3.95ILNGSPVDD1689 pKa = 3.83VNDD1692 pKa = 3.47INVTILTPAAHH1703 pKa = 6.7AGVVLDD1709 pKa = 4.1PATGIVSVTANVPDD1723 pKa = 3.48GTYY1726 pKa = 9.72IIEE1729 pKa = 4.03YY1730 pKa = 9.15QICEE1734 pKa = 4.25KK1735 pKa = 10.9LNPTNCKK1742 pKa = 7.83TATVTVVVEE1751 pKa = 4.28PPVSSVTGTVFTDD1764 pKa = 3.34VNGDD1768 pKa = 3.51GVLGPDD1774 pKa = 3.43EE1775 pKa = 4.35PRR1777 pKa = 11.84RR1778 pKa = 11.84AGWIVEE1784 pKa = 3.85IMKK1787 pKa = 10.62DD1788 pKa = 3.65GVVVATTTTDD1798 pKa = 2.84ANGDD1802 pKa = 3.71YY1803 pKa = 10.57RR1804 pKa = 11.84VDD1806 pKa = 3.7GLLSGPGYY1814 pKa = 10.57DD1815 pKa = 2.52IVFRR1819 pKa = 11.84NPEE1822 pKa = 3.94NNVVYY1827 pKa = 10.7DD1828 pKa = 3.99KK1829 pKa = 11.33IEE1831 pKa = 3.99GVNLVNNTVVIDD1843 pKa = 3.67QNQPIDD1849 pKa = 3.99PSGVIYY1855 pKa = 10.49DD1856 pKa = 4.32SITRR1860 pKa = 11.84NPISGVTVRR1869 pKa = 11.84LLGPDD1874 pKa = 4.09GNPLPSICFVDD1885 pKa = 4.83ASQASQTTGASGEE1898 pKa = 3.99YY1899 pKa = 9.91RR1900 pKa = 11.84FDD1902 pKa = 4.93IIPGAAPQCPASEE1915 pKa = 4.17TVYY1918 pKa = 9.94TIQVTPPVGFADD1930 pKa = 4.65GSTVLVPLPGPFDD1943 pKa = 4.12PSGLPAPVRR1952 pKa = 11.84ISPDD1956 pKa = 2.86ATPPQGSDD1964 pKa = 3.06TPFYY1968 pKa = 10.95LSFRR1972 pKa = 11.84LQPGDD1977 pKa = 3.53PDD1979 pKa = 3.65VVNNHH1984 pKa = 6.21IALDD1988 pKa = 3.79PFLNRR1993 pKa = 11.84TPLVVTKK2000 pKa = 9.67TSIKK2004 pKa = 10.17RR2005 pKa = 11.84SASTGDD2011 pKa = 3.5LVPYY2015 pKa = 9.85EE2016 pKa = 3.69ITVRR2020 pKa = 11.84NTEE2023 pKa = 3.71NAQRR2027 pKa = 11.84AGVDD2031 pKa = 3.55VVDD2034 pKa = 4.53ILPAGMKK2041 pKa = 9.94YY2042 pKa = 10.69VLGTASVDD2050 pKa = 3.49GVANEE2055 pKa = 5.13PIATNNNRR2063 pKa = 11.84EE2064 pKa = 4.18LRR2066 pKa = 11.84WTGQVIPANGSVRR2079 pKa = 11.84YY2080 pKa = 9.8NLTLVVGAGVTGGEE2094 pKa = 4.27KK2095 pKa = 10.79VNTGLAQNAADD2106 pKa = 5.45FSAISNRR2113 pKa = 11.84GTAVVAIVPSAVFDD2127 pKa = 4.06CSEE2130 pKa = 3.96LLGKK2134 pKa = 10.38VFEE2137 pKa = 4.63DD2138 pKa = 3.88RR2139 pKa = 11.84NRR2141 pKa = 11.84NGYY2144 pKa = 8.45QDD2146 pKa = 3.64EE2147 pKa = 4.53NEE2149 pKa = 3.99PGIAGVRR2156 pKa = 11.84LATVNGQLITTDD2168 pKa = 2.82EE2169 pKa = 4.39FGRR2172 pKa = 11.84YY2173 pKa = 8.91HH2174 pKa = 6.79IACAAVPDD2182 pKa = 4.07ARR2184 pKa = 11.84IGSNFVLKK2192 pKa = 10.67VDD2194 pKa = 3.83TRR2196 pKa = 11.84TLPIGWEE2203 pKa = 4.13VTFDD2207 pKa = 3.5NPKK2210 pKa = 10.32SIRR2213 pKa = 11.84LTRR2216 pKa = 11.84GKK2218 pKa = 10.41FGEE2221 pKa = 4.42LNFGVAPRR2229 pKa = 11.84EE2230 pKa = 3.87EE2231 pKa = 4.28GAPTNSNGKK2240 pKa = 10.14GEE2242 pKa = 4.05
MM1 pKa = 7.53RR2 pKa = 11.84HH3 pKa = 6.14LAFDD7 pKa = 3.56NQKK10 pKa = 8.29TVISSSEE17 pKa = 3.83QARR20 pKa = 11.84KK21 pKa = 9.25LLKK24 pKa = 9.34RR25 pKa = 11.84AKK27 pKa = 8.39WLAGASILAASSTAMAAPSLVVNNVGTPTVVGTGQGKK64 pKa = 9.02RR65 pKa = 11.84AIWTNAGTVGSDD77 pKa = 3.32TIDD80 pKa = 3.31LVGVITTATLNHH92 pKa = 6.18TWSTTANRR100 pKa = 11.84PSITSVGPDD109 pKa = 3.78DD110 pKa = 5.92IFIEE114 pKa = 3.97WRR116 pKa = 11.84LYY118 pKa = 10.42RR119 pKa = 11.84AGTYY123 pKa = 10.58NITTNSGGVPVVADD137 pKa = 2.84VHH139 pKa = 6.12VQFNDD144 pKa = 2.69VDD146 pKa = 3.86GPNNEE151 pKa = 4.33RR152 pKa = 11.84IYY154 pKa = 11.23LPVCQGDD161 pKa = 3.83ISWVRR166 pKa = 11.84IDD168 pKa = 3.49ATATTGRR175 pKa = 11.84AFGAVAGQAEE185 pKa = 4.88TFSLIGDD192 pKa = 3.56QNYY195 pKa = 10.22NNQPVSGLEE204 pKa = 3.95ALWKK208 pKa = 10.51DD209 pKa = 2.99RR210 pKa = 11.84STFTMGRR217 pKa = 11.84TANSGFLIRR226 pKa = 11.84FDD228 pKa = 3.77NPTYY232 pKa = 10.88SAFDD236 pKa = 3.57TLDD239 pKa = 3.52FEE241 pKa = 5.23CADD244 pKa = 4.22FKK246 pKa = 11.52PPVTVNDD253 pKa = 3.84SKK255 pKa = 11.45EE256 pKa = 4.25GVPGTSTIVDD266 pKa = 3.27ILDD269 pKa = 3.9NDD271 pKa = 4.16SSATLNNNPTNNNSLKK287 pKa = 10.55ASEE290 pKa = 4.23FARR293 pKa = 11.84ASVNLVPPPSATGIITDD310 pKa = 3.69SFGDD314 pKa = 3.63VIGFTVPGEE323 pKa = 4.51GVWSYY328 pKa = 12.09SDD330 pKa = 3.21TTGQLTFTPDD340 pKa = 3.3PSFVGYY346 pKa = 10.81ASTVNYY352 pKa = 9.32TVDD355 pKa = 3.26NALGIQSNQATVTVWYY371 pKa = 9.12PGIGVTKK378 pKa = 10.66ASTFNDD384 pKa = 3.76LNGDD388 pKa = 3.61GYY390 pKa = 10.93GQVGEE395 pKa = 4.53TINYY399 pKa = 7.86IYY401 pKa = 9.95QVKK404 pKa = 10.36SYY406 pKa = 10.57GAEE409 pKa = 3.93PLRR412 pKa = 11.84NPSLTEE418 pKa = 3.81TTFTGAGTPPTPTYY432 pKa = 11.2VSGDD436 pKa = 3.87TNNDD440 pKa = 2.99GYY442 pKa = 10.71IGLTEE447 pKa = 3.83TWFYY451 pKa = 10.73SAVYY455 pKa = 10.3SLVAGDD461 pKa = 4.36LSGTGVNNSATASGNTAAGTVVNDD485 pKa = 3.86VSDD488 pKa = 3.85SANPADD494 pKa = 4.3GNNNTKK500 pKa = 10.45SGPGPGNNDD509 pKa = 2.87ATVTAVPRR517 pKa = 11.84APIAASNDD525 pKa = 3.88TQSGTITNDD534 pKa = 2.97GQANAFNVLSNDD546 pKa = 3.61SLKK549 pKa = 11.01GAAPTPSNVAITVTTPASHH568 pKa = 7.13AGVTLNTSTGQVSVAPGTPAGNYY591 pKa = 6.86TINYY595 pKa = 6.67QICEE599 pKa = 3.9IGNPTNCANAVATVNVSLRR618 pKa = 11.84PINADD623 pKa = 3.43DD624 pKa = 4.91DD625 pKa = 4.5SGPSVNSTNGSASLYY640 pKa = 10.37NVLSNDD646 pKa = 3.5TFNGGAATTPNVTINVTSPATPAFAGASVPSLNISTGVVSVPAGTPAGTYY696 pKa = 9.9NIGYY700 pKa = 7.32EE701 pKa = 4.17ICDD704 pKa = 3.39SAVPTNCEE712 pKa = 3.33AAIATIIVTPTPIAASNDD730 pKa = 3.13SGPAVNGAAGNTNAFNAFTNDD751 pKa = 3.37SFNGNPVNLSLVTATVTSPATPLSAGAPVPSLNTATGVVSVPAGTPAGSYY801 pKa = 8.84TIGYY805 pKa = 7.73RR806 pKa = 11.84ICEE809 pKa = 4.05NANPTNCANASITVQVAASPIAANNDD835 pKa = 2.96STPAVNSANGGSNLVNALGNDD856 pKa = 4.04SLNGAAPTASNVNITVTTPASNAGVTMNTATGQVSVAPGTPAGNYY901 pKa = 9.64SIGYY905 pKa = 7.13QICEE909 pKa = 4.1KK910 pKa = 10.7LNPSNCAIANVTIPVNVTPIAASADD935 pKa = 3.69TPPAQNGRR943 pKa = 11.84TGANDD948 pKa = 2.91IVNAFTNDD956 pKa = 3.54SLNGSPVNVADD967 pKa = 3.91IEE969 pKa = 4.32ATITAPATPLTAGAPVPVMDD989 pKa = 5.18PATGLVDD996 pKa = 4.08VPAGTPAGTYY1006 pKa = 8.56TIAYY1010 pKa = 7.19EE1011 pKa = 4.12ICEE1014 pKa = 4.19KK1015 pKa = 10.45TNPTNCANSSVTVIVSAAPITALNDD1040 pKa = 3.25NAGTVTTANGGANLINALGNDD1061 pKa = 4.16TLNGSPVALADD1072 pKa = 3.64VNLTVTAPASNAGVTLNTATGQVSVAPGTPAGNYY1106 pKa = 9.64SIGYY1110 pKa = 7.13QICEE1114 pKa = 4.17KK1115 pKa = 10.75LNPTNCQSATIAVVVDD1131 pKa = 4.03VTPLNASNDD1140 pKa = 3.23SPAPVNGANGGNDD1153 pKa = 3.64IINAFANDD1161 pKa = 4.2TLNGAPVNVADD1172 pKa = 4.75ISATITAPATPLTAGAPVPVMDD1194 pKa = 5.18PATGLVDD1201 pKa = 4.08VPAGTPAGTYY1211 pKa = 8.56TIAYY1215 pKa = 7.18EE1216 pKa = 3.77ICEE1219 pKa = 4.08NSNPTNCKK1227 pKa = 7.55TATVQVVVTAAPITADD1243 pKa = 3.68ADD1245 pKa = 3.64SPAPVNGATGGNDD1258 pKa = 3.53IINAFANDD1266 pKa = 4.17SLNGAPVNVADD1277 pKa = 4.75IEE1279 pKa = 4.43ATVTSPAMPLTAGAPVPVMDD1299 pKa = 5.18PATGLVDD1306 pKa = 4.08VPAGTPAGTYY1316 pKa = 8.56TIAYY1320 pKa = 7.25EE1321 pKa = 4.01ICEE1324 pKa = 4.09KK1325 pKa = 10.94LNPNNCASSTVNVVVTAAPITADD1348 pKa = 3.68ADD1350 pKa = 3.64SPAPVNGATGGNDD1363 pKa = 3.53IINAFANDD1371 pKa = 4.17SLNGAPVNVADD1382 pKa = 4.75IEE1384 pKa = 4.43ATVTSPATPLTAGAPVPVMDD1404 pKa = 5.18PATGLVDD1411 pKa = 3.83VPVGTPAGTYY1421 pKa = 8.32TIAYY1425 pKa = 7.25EE1426 pKa = 4.01ICEE1429 pKa = 4.09KK1430 pKa = 10.94LNPNNCATNNVTVVVTAAPIAASNDD1455 pKa = 3.02AVGPVPGISGGSNLINALNNDD1476 pKa = 4.15MLNGASVAIADD1487 pKa = 3.74INVTVTSPASHH1498 pKa = 6.82AGVTLDD1504 pKa = 4.06PATGMVSVAPNTPAGTYY1521 pKa = 9.0TIGYY1525 pKa = 7.18QICEE1529 pKa = 4.21KK1530 pKa = 10.84LNPTNCANAIISVTVDD1546 pKa = 3.73AAPIMADD1553 pKa = 3.01VDD1555 pKa = 4.0SPAPVNGANGGNDD1568 pKa = 3.64IINAFANDD1576 pKa = 4.17SLNGAPVNVADD1587 pKa = 4.46IDD1589 pKa = 3.98ATITTPATPATPGAPVPVMDD1609 pKa = 5.18PATGLVDD1616 pKa = 3.54VSVGTPAGTYY1626 pKa = 8.32TIAYY1630 pKa = 7.51EE1631 pKa = 4.19ICQKK1635 pKa = 10.92LNPTNCASSNVTIVVEE1651 pKa = 4.14APEE1654 pKa = 3.83ILAANDD1660 pKa = 2.82VPAPVRR1666 pKa = 11.84SGVGNPNAINAFANDD1681 pKa = 3.95ILNGSPVDD1689 pKa = 3.83VNDD1692 pKa = 3.47INVTILTPAAHH1703 pKa = 6.7AGVVLDD1709 pKa = 4.1PATGIVSVTANVPDD1723 pKa = 3.48GTYY1726 pKa = 9.72IIEE1729 pKa = 4.03YY1730 pKa = 9.15QICEE1734 pKa = 4.25KK1735 pKa = 10.9LNPTNCKK1742 pKa = 7.83TATVTVVVEE1751 pKa = 4.28PPVSSVTGTVFTDD1764 pKa = 3.34VNGDD1768 pKa = 3.51GVLGPDD1774 pKa = 3.43EE1775 pKa = 4.35PRR1777 pKa = 11.84RR1778 pKa = 11.84AGWIVEE1784 pKa = 3.85IMKK1787 pKa = 10.62DD1788 pKa = 3.65GVVVATTTTDD1798 pKa = 2.84ANGDD1802 pKa = 3.71YY1803 pKa = 10.57RR1804 pKa = 11.84VDD1806 pKa = 3.7GLLSGPGYY1814 pKa = 10.57DD1815 pKa = 2.52IVFRR1819 pKa = 11.84NPEE1822 pKa = 3.94NNVVYY1827 pKa = 10.7DD1828 pKa = 3.99KK1829 pKa = 11.33IEE1831 pKa = 3.99GVNLVNNTVVIDD1843 pKa = 3.67QNQPIDD1849 pKa = 3.99PSGVIYY1855 pKa = 10.49DD1856 pKa = 4.32SITRR1860 pKa = 11.84NPISGVTVRR1869 pKa = 11.84LLGPDD1874 pKa = 4.09GNPLPSICFVDD1885 pKa = 4.83ASQASQTTGASGEE1898 pKa = 3.99YY1899 pKa = 9.91RR1900 pKa = 11.84FDD1902 pKa = 4.93IIPGAAPQCPASEE1915 pKa = 4.17TVYY1918 pKa = 9.94TIQVTPPVGFADD1930 pKa = 4.65GSTVLVPLPGPFDD1943 pKa = 4.12PSGLPAPVRR1952 pKa = 11.84ISPDD1956 pKa = 2.86ATPPQGSDD1964 pKa = 3.06TPFYY1968 pKa = 10.95LSFRR1972 pKa = 11.84LQPGDD1977 pKa = 3.53PDD1979 pKa = 3.65VVNNHH1984 pKa = 6.21IALDD1988 pKa = 3.79PFLNRR1993 pKa = 11.84TPLVVTKK2000 pKa = 9.67TSIKK2004 pKa = 10.17RR2005 pKa = 11.84SASTGDD2011 pKa = 3.5LVPYY2015 pKa = 9.85EE2016 pKa = 3.69ITVRR2020 pKa = 11.84NTEE2023 pKa = 3.71NAQRR2027 pKa = 11.84AGVDD2031 pKa = 3.55VVDD2034 pKa = 4.53ILPAGMKK2041 pKa = 9.94YY2042 pKa = 10.69VLGTASVDD2050 pKa = 3.49GVANEE2055 pKa = 5.13PIATNNNRR2063 pKa = 11.84EE2064 pKa = 4.18LRR2066 pKa = 11.84WTGQVIPANGSVRR2079 pKa = 11.84YY2080 pKa = 9.8NLTLVVGAGVTGGEE2094 pKa = 4.27KK2095 pKa = 10.79VNTGLAQNAADD2106 pKa = 5.45FSAISNRR2113 pKa = 11.84GTAVVAIVPSAVFDD2127 pKa = 4.06CSEE2130 pKa = 3.96LLGKK2134 pKa = 10.38VFEE2137 pKa = 4.63DD2138 pKa = 3.88RR2139 pKa = 11.84NRR2141 pKa = 11.84NGYY2144 pKa = 8.45QDD2146 pKa = 3.64EE2147 pKa = 4.53NEE2149 pKa = 3.99PGIAGVRR2156 pKa = 11.84LATVNGQLITTDD2168 pKa = 2.82EE2169 pKa = 4.39FGRR2172 pKa = 11.84YY2173 pKa = 8.91HH2174 pKa = 6.79IACAAVPDD2182 pKa = 4.07ARR2184 pKa = 11.84IGSNFVLKK2192 pKa = 10.67VDD2194 pKa = 3.83TRR2196 pKa = 11.84TLPIGWEE2203 pKa = 4.13VTFDD2207 pKa = 3.5NPKK2210 pKa = 10.32SIRR2213 pKa = 11.84LTRR2216 pKa = 11.84GKK2218 pKa = 10.41FGEE2221 pKa = 4.42LNFGVAPRR2229 pKa = 11.84EE2230 pKa = 3.87EE2231 pKa = 4.28GAPTNSNGKK2240 pKa = 10.14GEE2242 pKa = 4.05
Molecular weight: 227.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3JAU6|A0A1L3JAU6_9SPHN Alcohol dehydrogenase OS=Sphingopyxis sp. LPB0140 OX=1913578 GN=LPB140_04890 PE=3 SV=1
MM1 pKa = 7.64KK2 pKa = 10.54LNLGLFSLLILAMLAALFAGKK23 pKa = 9.43IWVPVDD29 pKa = 3.67EE30 pKa = 4.87IFGGGRR36 pKa = 11.84GALIIAEE43 pKa = 4.28LRR45 pKa = 11.84TPRR48 pKa = 11.84IILGVIIGGALGLSGAVMQGYY69 pKa = 9.33LRR71 pKa = 11.84NPLADD76 pKa = 4.25PGLFGVSSGAALGAVIAIFWGLAGEE101 pKa = 4.73IWALPLFALIGAGGSMALLGMLAGRR126 pKa = 11.84SGSLILITLGGVMLSTLTGSLTALVISLAPTPFATSQIITWLMGALSDD174 pKa = 4.68RR175 pKa = 11.84SWSDD179 pKa = 3.05VYY181 pKa = 10.72IAAPLIFIGMMILFATARR199 pKa = 11.84ALDD202 pKa = 3.93VLTLGDD208 pKa = 3.65VVARR212 pKa = 11.84SMGVDD217 pKa = 3.28MVRR220 pKa = 11.84LQWQVILGTGICVGAAVAAAGVIGFVGLMVPHH252 pKa = 7.89IIRR255 pKa = 11.84PFVGHH260 pKa = 6.77RR261 pKa = 11.84PSALLLPSALAGAILIISSDD281 pKa = 3.34ALVRR285 pKa = 11.84ILPTVSEE292 pKa = 4.36LRR294 pKa = 11.84LGIAMTMLGTPFFLFLLFKK313 pKa = 9.51MRR315 pKa = 11.84RR316 pKa = 11.84GIMM319 pKa = 3.43
MM1 pKa = 7.64KK2 pKa = 10.54LNLGLFSLLILAMLAALFAGKK23 pKa = 9.43IWVPVDD29 pKa = 3.67EE30 pKa = 4.87IFGGGRR36 pKa = 11.84GALIIAEE43 pKa = 4.28LRR45 pKa = 11.84TPRR48 pKa = 11.84IILGVIIGGALGLSGAVMQGYY69 pKa = 9.33LRR71 pKa = 11.84NPLADD76 pKa = 4.25PGLFGVSSGAALGAVIAIFWGLAGEE101 pKa = 4.73IWALPLFALIGAGGSMALLGMLAGRR126 pKa = 11.84SGSLILITLGGVMLSTLTGSLTALVISLAPTPFATSQIITWLMGALSDD174 pKa = 4.68RR175 pKa = 11.84SWSDD179 pKa = 3.05VYY181 pKa = 10.72IAAPLIFIGMMILFATARR199 pKa = 11.84ALDD202 pKa = 3.93VLTLGDD208 pKa = 3.65VVARR212 pKa = 11.84SMGVDD217 pKa = 3.28MVRR220 pKa = 11.84LQWQVILGTGICVGAAVAAAGVIGFVGLMVPHH252 pKa = 7.89IIRR255 pKa = 11.84PFVGHH260 pKa = 6.77RR261 pKa = 11.84PSALLLPSALAGAILIISSDD281 pKa = 3.34ALVRR285 pKa = 11.84ILPTVSEE292 pKa = 4.36LRR294 pKa = 11.84LGIAMTMLGTPFFLFLLFKK313 pKa = 9.51MRR315 pKa = 11.84RR316 pKa = 11.84GIMM319 pKa = 3.43
Molecular weight: 33.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
760127 |
53 |
3009 |
323.5 |
35.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.665 ± 0.055 | 0.874 ± 0.017 |
6.091 ± 0.043 | 5.538 ± 0.049 |
3.971 ± 0.033 | 8.107 ± 0.045 |
2.083 ± 0.029 | 7.347 ± 0.048 |
5.132 ± 0.044 | 9.037 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.008 ± 0.03 | 4.426 ± 0.047 |
4.445 ± 0.032 | 3.607 ± 0.032 |
5.177 ± 0.044 | 6.03 ± 0.046 |
4.555 ± 0.045 | 5.896 ± 0.057 |
1.302 ± 0.019 | 2.709 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
