
Alteromonas aestuariivivens
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Alteromonas
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
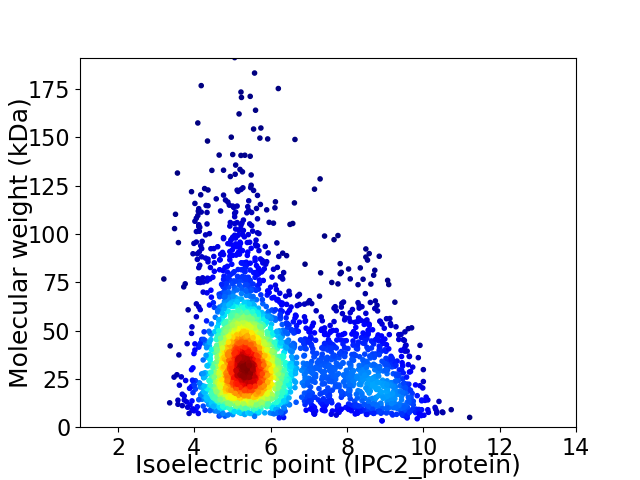
Virtual 2D-PAGE plot for 3306 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3D8M4I8|A0A3D8M4I8_9ALTE Na+/H+ antiporter subunit C OS=Alteromonas aestuariivivens OX=1938339 GN=DXV75_13275 PE=4 SV=1
NN1 pKa = 7.69GSLTLNSDD9 pKa = 3.41GTFTYY14 pKa = 10.66VHH16 pKa = 7.38DD17 pKa = 4.7GGEE20 pKa = 4.29TTSDD24 pKa = 3.1SFTYY28 pKa = 9.8QANDD32 pKa = 3.42GSSTSNTVTVSISITADD49 pKa = 2.57NDD51 pKa = 3.57APVAVADD58 pKa = 4.28SYY60 pKa = 11.9SVTEE64 pKa = 4.23GGTLNGTSVLSNDD77 pKa = 3.22TDD79 pKa = 3.85AEE81 pKa = 4.42SDD83 pKa = 3.66SLTAVQVSSPANGSLTLNSDD103 pKa = 3.41GTFTYY108 pKa = 10.66VHH110 pKa = 7.38DD111 pKa = 4.7GGEE114 pKa = 4.29TTSDD118 pKa = 3.1SFTYY122 pKa = 9.8QANDD126 pKa = 3.35GSSNSNTVTVSISITADD143 pKa = 2.63NDD145 pKa = 3.53APVAKK150 pKa = 9.48DD151 pKa = 3.29DD152 pKa = 4.13SVAVDD157 pKa = 4.21EE158 pKa = 5.74DD159 pKa = 3.74STDD162 pKa = 3.29NVIPVLDD169 pKa = 3.97NDD171 pKa = 4.05IDD173 pKa = 4.09PEE175 pKa = 4.48GDD177 pKa = 3.32EE178 pKa = 4.36LTITQATASEE188 pKa = 4.65GSVSINSDD196 pKa = 2.9GTLSYY201 pKa = 10.56TPEE204 pKa = 3.84EE205 pKa = 4.14NATDD209 pKa = 3.78SVTIRR214 pKa = 11.84YY215 pKa = 7.11TISDD219 pKa = 4.06ANGLTDD225 pKa = 3.58TASVTVSINPQNDD238 pKa = 3.1APQADD243 pKa = 4.2AQTITLDD250 pKa = 3.26EE251 pKa = 4.45DD252 pKa = 4.33TNTGITLTATDD263 pKa = 3.94IDD265 pKa = 4.46GDD267 pKa = 4.15SLSYY271 pKa = 10.8EE272 pKa = 3.96LVSQPEE278 pKa = 4.16NGTLTGNAPEE288 pKa = 4.6LVYY291 pKa = 10.68TPAPDD296 pKa = 4.88FFGQDD301 pKa = 2.7EE302 pKa = 4.67FTFRR306 pKa = 11.84ANDD309 pKa = 3.7GTEE312 pKa = 3.87DD313 pKa = 3.69SEE315 pKa = 4.73LAVISITVEE324 pKa = 3.75DD325 pKa = 4.06VLDD328 pKa = 4.1NKK330 pKa = 10.96APLAVNDD337 pKa = 3.8AFEE340 pKa = 4.12VLKK343 pKa = 11.21NSTANRR349 pKa = 11.84LDD351 pKa = 3.36VLLNDD356 pKa = 4.82SDD358 pKa = 4.54PDD360 pKa = 3.76GDD362 pKa = 4.48KK363 pKa = 9.86ITVVQVYY370 pKa = 10.54ANLGEE375 pKa = 4.28VSVTTDD381 pKa = 3.01GAVVYY386 pKa = 10.9SPMTDD391 pKa = 2.76STQNDD396 pKa = 3.85LLTYY400 pKa = 10.3VIEE403 pKa = 4.68DD404 pKa = 3.67EE405 pKa = 4.61FGEE408 pKa = 4.4SASAEE413 pKa = 3.89VHH415 pKa = 5.02ITVVTEE421 pKa = 4.38DD422 pKa = 4.2NLPPIAVDD430 pKa = 4.89DD431 pKa = 4.37SVTMQTGEE439 pKa = 4.38TITLDD444 pKa = 3.57VLANDD449 pKa = 4.59YY450 pKa = 11.21DD451 pKa = 4.35PEE453 pKa = 6.49DD454 pKa = 3.9DD455 pKa = 5.28AIEE458 pKa = 4.04LVAVSGDD465 pKa = 3.28VGEE468 pKa = 4.28VVIVDD473 pKa = 4.04GALQFTPQSNIHH485 pKa = 6.33GDD487 pKa = 3.6YY488 pKa = 10.77LITYY492 pKa = 7.36SIRR495 pKa = 11.84DD496 pKa = 3.37AAGNFASAEE505 pKa = 3.98VLVNVEE511 pKa = 4.34SEE513 pKa = 4.17SGPFITLPEE522 pKa = 4.49DD523 pKa = 3.36LCSEE527 pKa = 4.32LTVMANALYY536 pKa = 9.99TKK538 pKa = 10.45VDD540 pKa = 4.04LGQASAVDD548 pKa = 3.35RR549 pKa = 11.84FGNVLPVSLLDD560 pKa = 4.33GNTLYY565 pKa = 10.54PPGINQAFWSATDD578 pKa = 3.71AEE580 pKa = 5.13GNTAIATQLVCVAPLVSIQKK600 pKa = 10.0DD601 pKa = 3.32QTVLKK606 pKa = 10.44GSSTTIGVYY615 pKa = 10.74LNGEE619 pKa = 4.26SPVYY623 pKa = 9.85PLVVPYY629 pKa = 9.48TLSGDD634 pKa = 3.64ATEE637 pKa = 4.71EE638 pKa = 4.03DD639 pKa = 4.28HH640 pKa = 7.48NLEE643 pKa = 4.51AGDD646 pKa = 4.93LIIEE650 pKa = 4.66SGTTAYY656 pKa = 9.69IALDD660 pKa = 3.49TFINEE665 pKa = 4.27SNRR668 pKa = 11.84SDD670 pKa = 3.37RR671 pKa = 11.84VVTVSLDD678 pKa = 3.19EE679 pKa = 4.2SLNRR683 pKa = 11.84GAKK686 pKa = 9.89DD687 pKa = 3.2EE688 pKa = 4.76HH689 pKa = 6.41NVLLTEE695 pKa = 4.44RR696 pKa = 11.84NIEE699 pKa = 4.02PDD701 pKa = 2.85VTLQVLQNHH710 pKa = 6.58IKK712 pKa = 10.4RR713 pKa = 11.84LTVSRR718 pKa = 11.84RR719 pKa = 11.84GGAISVISMVEE730 pKa = 4.05DD731 pKa = 3.89PNLTDD736 pKa = 2.97SHH738 pKa = 6.98QYY740 pKa = 10.23AWITSDD746 pKa = 2.87HH747 pKa = 6.44RR748 pKa = 11.84LVNEE752 pKa = 4.19ATNDD756 pKa = 3.28TAFIFDD762 pKa = 4.31PSLLEE767 pKa = 3.76AGIYY771 pKa = 9.36HH772 pKa = 6.19VSIEE776 pKa = 4.38VTDD779 pKa = 3.63SGAPQLSDD787 pKa = 3.48SEE789 pKa = 4.58TIYY792 pKa = 10.69IEE794 pKa = 4.18VVEE797 pKa = 4.54EE798 pKa = 3.99LLSFDD803 pKa = 4.58DD804 pKa = 5.39HH805 pKa = 8.93SLDD808 pKa = 4.36SDD810 pKa = 3.79GDD812 pKa = 4.31LIPDD816 pKa = 3.68HH817 pKa = 6.56MEE819 pKa = 4.85GYY821 pKa = 10.42QDD823 pKa = 3.56TDD825 pKa = 3.08GDD827 pKa = 4.59GIPDD831 pKa = 3.55FLDD834 pKa = 5.67RR835 pKa = 11.84IDD837 pKa = 3.84EE838 pKa = 4.49CNVLQEE844 pKa = 4.13VAKK847 pKa = 10.9VHH849 pKa = 6.77DD850 pKa = 4.36AFLIEE855 pKa = 4.34GQAGVCLRR863 pKa = 11.84RR864 pKa = 11.84GDD866 pKa = 3.76HH867 pKa = 6.77SIGGEE872 pKa = 3.92TGGAQITNDD881 pKa = 5.07DD882 pKa = 3.76IAQDD886 pKa = 3.8EE887 pKa = 4.5LDD889 pKa = 4.51EE890 pKa = 5.64LVDD893 pKa = 5.54DD894 pKa = 5.03PDD896 pKa = 5.75AINVGGIFDD905 pKa = 4.33YY906 pKa = 10.55IAYY909 pKa = 9.81GMPEE913 pKa = 4.03QGTQFAIVMPQRR925 pKa = 11.84KK926 pKa = 8.56PIPADD931 pKa = 2.89AVYY934 pKa = 10.59RR935 pKa = 11.84KK936 pKa = 9.05FRR938 pKa = 11.84PGSGWGFFIEE948 pKa = 5.17DD949 pKa = 4.22ANNSLWSTPGDD960 pKa = 3.4VGFCPPPDD968 pKa = 3.69VNSGNSVWTPGLTEE982 pKa = 4.09GHH984 pKa = 6.56WCVQQIIEE992 pKa = 4.66DD993 pKa = 3.92GGVNDD998 pKa = 6.0DD999 pKa = 4.58DD1000 pKa = 4.71NLVNGTIVDD1009 pKa = 3.85PGGVGVMITSNALPEE1024 pKa = 4.47ASDD1027 pKa = 4.05DD1028 pKa = 4.2SVTLVVNQEE1037 pKa = 3.59ITVDD1041 pKa = 4.03VIANDD1046 pKa = 3.6TDD1048 pKa = 3.81QNGDD1052 pKa = 3.79PLLLTSATTTIGEE1065 pKa = 4.09VSIVDD1070 pKa = 3.6NAIHH1074 pKa = 5.96YY1075 pKa = 8.3VAALDD1080 pKa = 3.84YY1081 pKa = 11.12VGEE1084 pKa = 3.95ITINYY1089 pKa = 8.35GVTDD1093 pKa = 3.94SNGGTDD1099 pKa = 3.77HH1100 pKa = 7.47AVLTINLIDD1109 pKa = 3.93NKK1111 pKa = 10.91APLITNDD1118 pKa = 3.45SSEE1121 pKa = 4.06ILQGEE1126 pKa = 4.43TATLNLLTNDD1136 pKa = 4.13SDD1138 pKa = 4.41PDD1140 pKa = 3.88GDD1142 pKa = 3.87QMRR1145 pKa = 11.84LLSVEE1150 pKa = 3.86HH1151 pKa = 6.56SGVSFSEE1158 pKa = 3.84NGEE1161 pKa = 4.42VVFTPQSDD1169 pKa = 4.17FYY1171 pKa = 10.95GVLVIDD1177 pKa = 4.16YY1178 pKa = 9.42QVSDD1182 pKa = 2.95EE1183 pKa = 4.5WGNQSFGQWSVTVTEE1198 pKa = 3.83YY1199 pKa = 11.03HH1200 pKa = 6.92RR1201 pKa = 11.84IEE1203 pKa = 4.23ASTRR1207 pKa = 11.84GGGSLYY1213 pKa = 11.0GLLLLGVFVSVIRR1226 pKa = 11.84KK1227 pKa = 8.61VNWKK1231 pKa = 10.1ANRR1234 pKa = 11.84GGII1237 pKa = 3.85
NN1 pKa = 7.69GSLTLNSDD9 pKa = 3.41GTFTYY14 pKa = 10.66VHH16 pKa = 7.38DD17 pKa = 4.7GGEE20 pKa = 4.29TTSDD24 pKa = 3.1SFTYY28 pKa = 9.8QANDD32 pKa = 3.42GSSTSNTVTVSISITADD49 pKa = 2.57NDD51 pKa = 3.57APVAVADD58 pKa = 4.28SYY60 pKa = 11.9SVTEE64 pKa = 4.23GGTLNGTSVLSNDD77 pKa = 3.22TDD79 pKa = 3.85AEE81 pKa = 4.42SDD83 pKa = 3.66SLTAVQVSSPANGSLTLNSDD103 pKa = 3.41GTFTYY108 pKa = 10.66VHH110 pKa = 7.38DD111 pKa = 4.7GGEE114 pKa = 4.29TTSDD118 pKa = 3.1SFTYY122 pKa = 9.8QANDD126 pKa = 3.35GSSNSNTVTVSISITADD143 pKa = 2.63NDD145 pKa = 3.53APVAKK150 pKa = 9.48DD151 pKa = 3.29DD152 pKa = 4.13SVAVDD157 pKa = 4.21EE158 pKa = 5.74DD159 pKa = 3.74STDD162 pKa = 3.29NVIPVLDD169 pKa = 3.97NDD171 pKa = 4.05IDD173 pKa = 4.09PEE175 pKa = 4.48GDD177 pKa = 3.32EE178 pKa = 4.36LTITQATASEE188 pKa = 4.65GSVSINSDD196 pKa = 2.9GTLSYY201 pKa = 10.56TPEE204 pKa = 3.84EE205 pKa = 4.14NATDD209 pKa = 3.78SVTIRR214 pKa = 11.84YY215 pKa = 7.11TISDD219 pKa = 4.06ANGLTDD225 pKa = 3.58TASVTVSINPQNDD238 pKa = 3.1APQADD243 pKa = 4.2AQTITLDD250 pKa = 3.26EE251 pKa = 4.45DD252 pKa = 4.33TNTGITLTATDD263 pKa = 3.94IDD265 pKa = 4.46GDD267 pKa = 4.15SLSYY271 pKa = 10.8EE272 pKa = 3.96LVSQPEE278 pKa = 4.16NGTLTGNAPEE288 pKa = 4.6LVYY291 pKa = 10.68TPAPDD296 pKa = 4.88FFGQDD301 pKa = 2.7EE302 pKa = 4.67FTFRR306 pKa = 11.84ANDD309 pKa = 3.7GTEE312 pKa = 3.87DD313 pKa = 3.69SEE315 pKa = 4.73LAVISITVEE324 pKa = 3.75DD325 pKa = 4.06VLDD328 pKa = 4.1NKK330 pKa = 10.96APLAVNDD337 pKa = 3.8AFEE340 pKa = 4.12VLKK343 pKa = 11.21NSTANRR349 pKa = 11.84LDD351 pKa = 3.36VLLNDD356 pKa = 4.82SDD358 pKa = 4.54PDD360 pKa = 3.76GDD362 pKa = 4.48KK363 pKa = 9.86ITVVQVYY370 pKa = 10.54ANLGEE375 pKa = 4.28VSVTTDD381 pKa = 3.01GAVVYY386 pKa = 10.9SPMTDD391 pKa = 2.76STQNDD396 pKa = 3.85LLTYY400 pKa = 10.3VIEE403 pKa = 4.68DD404 pKa = 3.67EE405 pKa = 4.61FGEE408 pKa = 4.4SASAEE413 pKa = 3.89VHH415 pKa = 5.02ITVVTEE421 pKa = 4.38DD422 pKa = 4.2NLPPIAVDD430 pKa = 4.89DD431 pKa = 4.37SVTMQTGEE439 pKa = 4.38TITLDD444 pKa = 3.57VLANDD449 pKa = 4.59YY450 pKa = 11.21DD451 pKa = 4.35PEE453 pKa = 6.49DD454 pKa = 3.9DD455 pKa = 5.28AIEE458 pKa = 4.04LVAVSGDD465 pKa = 3.28VGEE468 pKa = 4.28VVIVDD473 pKa = 4.04GALQFTPQSNIHH485 pKa = 6.33GDD487 pKa = 3.6YY488 pKa = 10.77LITYY492 pKa = 7.36SIRR495 pKa = 11.84DD496 pKa = 3.37AAGNFASAEE505 pKa = 3.98VLVNVEE511 pKa = 4.34SEE513 pKa = 4.17SGPFITLPEE522 pKa = 4.49DD523 pKa = 3.36LCSEE527 pKa = 4.32LTVMANALYY536 pKa = 9.99TKK538 pKa = 10.45VDD540 pKa = 4.04LGQASAVDD548 pKa = 3.35RR549 pKa = 11.84FGNVLPVSLLDD560 pKa = 4.33GNTLYY565 pKa = 10.54PPGINQAFWSATDD578 pKa = 3.71AEE580 pKa = 5.13GNTAIATQLVCVAPLVSIQKK600 pKa = 10.0DD601 pKa = 3.32QTVLKK606 pKa = 10.44GSSTTIGVYY615 pKa = 10.74LNGEE619 pKa = 4.26SPVYY623 pKa = 9.85PLVVPYY629 pKa = 9.48TLSGDD634 pKa = 3.64ATEE637 pKa = 4.71EE638 pKa = 4.03DD639 pKa = 4.28HH640 pKa = 7.48NLEE643 pKa = 4.51AGDD646 pKa = 4.93LIIEE650 pKa = 4.66SGTTAYY656 pKa = 9.69IALDD660 pKa = 3.49TFINEE665 pKa = 4.27SNRR668 pKa = 11.84SDD670 pKa = 3.37RR671 pKa = 11.84VVTVSLDD678 pKa = 3.19EE679 pKa = 4.2SLNRR683 pKa = 11.84GAKK686 pKa = 9.89DD687 pKa = 3.2EE688 pKa = 4.76HH689 pKa = 6.41NVLLTEE695 pKa = 4.44RR696 pKa = 11.84NIEE699 pKa = 4.02PDD701 pKa = 2.85VTLQVLQNHH710 pKa = 6.58IKK712 pKa = 10.4RR713 pKa = 11.84LTVSRR718 pKa = 11.84RR719 pKa = 11.84GGAISVISMVEE730 pKa = 4.05DD731 pKa = 3.89PNLTDD736 pKa = 2.97SHH738 pKa = 6.98QYY740 pKa = 10.23AWITSDD746 pKa = 2.87HH747 pKa = 6.44RR748 pKa = 11.84LVNEE752 pKa = 4.19ATNDD756 pKa = 3.28TAFIFDD762 pKa = 4.31PSLLEE767 pKa = 3.76AGIYY771 pKa = 9.36HH772 pKa = 6.19VSIEE776 pKa = 4.38VTDD779 pKa = 3.63SGAPQLSDD787 pKa = 3.48SEE789 pKa = 4.58TIYY792 pKa = 10.69IEE794 pKa = 4.18VVEE797 pKa = 4.54EE798 pKa = 3.99LLSFDD803 pKa = 4.58DD804 pKa = 5.39HH805 pKa = 8.93SLDD808 pKa = 4.36SDD810 pKa = 3.79GDD812 pKa = 4.31LIPDD816 pKa = 3.68HH817 pKa = 6.56MEE819 pKa = 4.85GYY821 pKa = 10.42QDD823 pKa = 3.56TDD825 pKa = 3.08GDD827 pKa = 4.59GIPDD831 pKa = 3.55FLDD834 pKa = 5.67RR835 pKa = 11.84IDD837 pKa = 3.84EE838 pKa = 4.49CNVLQEE844 pKa = 4.13VAKK847 pKa = 10.9VHH849 pKa = 6.77DD850 pKa = 4.36AFLIEE855 pKa = 4.34GQAGVCLRR863 pKa = 11.84RR864 pKa = 11.84GDD866 pKa = 3.76HH867 pKa = 6.77SIGGEE872 pKa = 3.92TGGAQITNDD881 pKa = 5.07DD882 pKa = 3.76IAQDD886 pKa = 3.8EE887 pKa = 4.5LDD889 pKa = 4.51EE890 pKa = 5.64LVDD893 pKa = 5.54DD894 pKa = 5.03PDD896 pKa = 5.75AINVGGIFDD905 pKa = 4.33YY906 pKa = 10.55IAYY909 pKa = 9.81GMPEE913 pKa = 4.03QGTQFAIVMPQRR925 pKa = 11.84KK926 pKa = 8.56PIPADD931 pKa = 2.89AVYY934 pKa = 10.59RR935 pKa = 11.84KK936 pKa = 9.05FRR938 pKa = 11.84PGSGWGFFIEE948 pKa = 5.17DD949 pKa = 4.22ANNSLWSTPGDD960 pKa = 3.4VGFCPPPDD968 pKa = 3.69VNSGNSVWTPGLTEE982 pKa = 4.09GHH984 pKa = 6.56WCVQQIIEE992 pKa = 4.66DD993 pKa = 3.92GGVNDD998 pKa = 6.0DD999 pKa = 4.58DD1000 pKa = 4.71NLVNGTIVDD1009 pKa = 3.85PGGVGVMITSNALPEE1024 pKa = 4.47ASDD1027 pKa = 4.05DD1028 pKa = 4.2SVTLVVNQEE1037 pKa = 3.59ITVDD1041 pKa = 4.03VIANDD1046 pKa = 3.6TDD1048 pKa = 3.81QNGDD1052 pKa = 3.79PLLLTSATTTIGEE1065 pKa = 4.09VSIVDD1070 pKa = 3.6NAIHH1074 pKa = 5.96YY1075 pKa = 8.3VAALDD1080 pKa = 3.84YY1081 pKa = 11.12VGEE1084 pKa = 3.95ITINYY1089 pKa = 8.35GVTDD1093 pKa = 3.94SNGGTDD1099 pKa = 3.77HH1100 pKa = 7.47AVLTINLIDD1109 pKa = 3.93NKK1111 pKa = 10.91APLITNDD1118 pKa = 3.45SSEE1121 pKa = 4.06ILQGEE1126 pKa = 4.43TATLNLLTNDD1136 pKa = 4.13SDD1138 pKa = 4.41PDD1140 pKa = 3.88GDD1142 pKa = 3.87QMRR1145 pKa = 11.84LLSVEE1150 pKa = 3.86HH1151 pKa = 6.56SGVSFSEE1158 pKa = 3.84NGEE1161 pKa = 4.42VVFTPQSDD1169 pKa = 4.17FYY1171 pKa = 10.95GVLVIDD1177 pKa = 4.16YY1178 pKa = 9.42QVSDD1182 pKa = 2.95EE1183 pKa = 4.5WGNQSFGQWSVTVTEE1198 pKa = 3.83YY1199 pKa = 11.03HH1200 pKa = 6.92RR1201 pKa = 11.84IEE1203 pKa = 4.23ASTRR1207 pKa = 11.84GGGSLYY1213 pKa = 11.0GLLLLGVFVSVIRR1226 pKa = 11.84KK1227 pKa = 8.61VNWKK1231 pKa = 10.1ANRR1234 pKa = 11.84GGII1237 pKa = 3.85
Molecular weight: 131.5 kDa
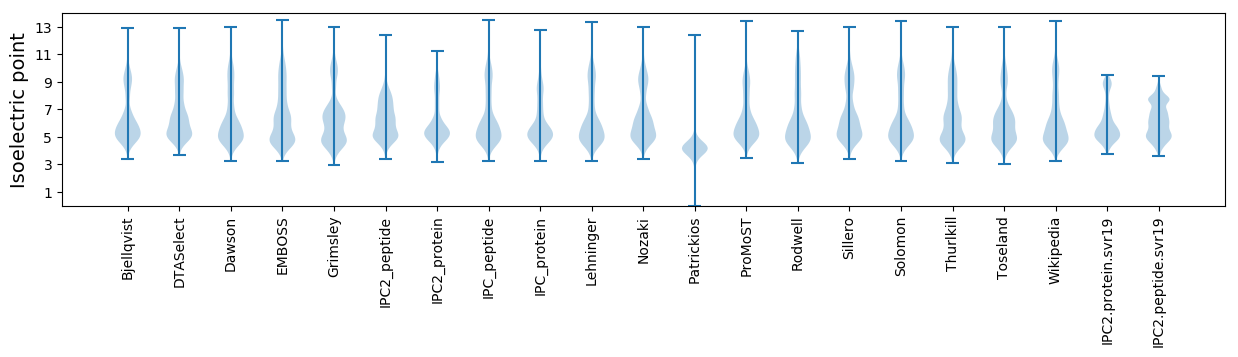
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3D8M6J8|A0A3D8M6J8_9ALTE ThuA domain-containing protein OS=Alteromonas aestuariivivens OX=1938339 GN=DXV75_11660 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1141386 |
28 |
1682 |
345.2 |
38.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.099 ± 0.045 | 1.021 ± 0.015 |
5.574 ± 0.037 | 6.12 ± 0.039 |
4.137 ± 0.025 | 7.09 ± 0.037 |
2.319 ± 0.022 | 5.624 ± 0.031 |
4.376 ± 0.034 | 10.382 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.507 ± 0.018 | 4.051 ± 0.031 |
4.298 ± 0.027 | 4.875 ± 0.038 |
5.186 ± 0.028 | 6.542 ± 0.03 |
5.19 ± 0.027 | 7.089 ± 0.037 |
1.362 ± 0.015 | 3.157 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
