
Idotea virus IWaV278
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses; Cruciviridae
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
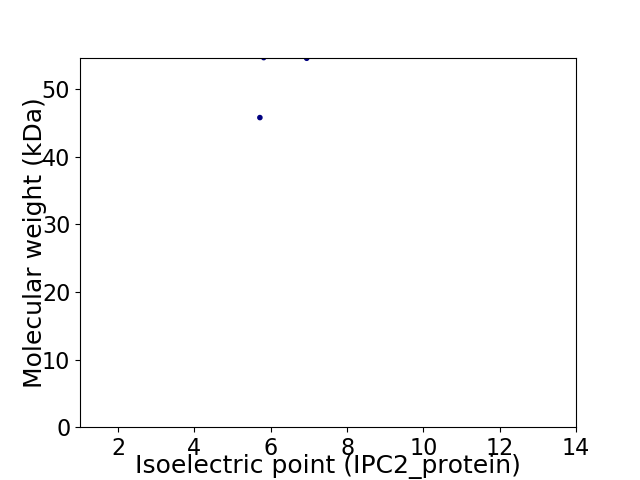
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4YQ33|A0A2H4YQ33_9VIRU Putative capsid protein OS=Idotea virus IWaV278 OX=2058759 PE=3 SV=1
MM1 pKa = 7.8PNGSKK6 pKa = 10.39RR7 pKa = 11.84FCFTHH12 pKa = 6.69FGCPDD17 pKa = 3.45EE18 pKa = 4.47FTNILQARR26 pKa = 11.84SPLCTKK32 pKa = 10.02WIVQQEE38 pKa = 4.05RR39 pKa = 11.84GQAEE43 pKa = 4.18GKK45 pKa = 10.69APLEE49 pKa = 4.28GANAQNGGLHH59 pKa = 5.58LQGYY63 pKa = 8.22IHH65 pKa = 6.8TKK67 pKa = 9.58RR68 pKa = 11.84VYY70 pKa = 10.13TFAQFLRR77 pKa = 11.84IWPVSHH83 pKa = 7.09VEE85 pKa = 3.79AAQGTEE91 pKa = 3.73AQNYY95 pKa = 7.44LYY97 pKa = 10.39CRR99 pKa = 11.84KK100 pKa = 10.2NEE102 pKa = 4.13TFTGKK107 pKa = 10.14YY108 pKa = 8.15RR109 pKa = 11.84LAMEE113 pKa = 4.65GSVWSRR119 pKa = 11.84DD120 pKa = 3.75VQRR123 pKa = 11.84LQAKK127 pKa = 8.34SRR129 pKa = 11.84KK130 pKa = 8.39DD131 pKa = 3.46AEE133 pKa = 4.06MQAIVDD139 pKa = 4.45KK140 pKa = 10.39IKK142 pKa = 10.88QGATTADD149 pKa = 3.02IARR152 pKa = 11.84NNTAYY157 pKa = 10.58FIRR160 pKa = 11.84HH161 pKa = 6.12CNGIPAAVRR170 pKa = 11.84HH171 pKa = 5.81HH172 pKa = 6.3QPAAPATRR180 pKa = 11.84PVHH183 pKa = 6.42VILLKK188 pKa = 10.93GSTGTGKK195 pKa = 10.6SYY197 pKa = 9.83WARR200 pKa = 11.84QLATYY205 pKa = 7.99QRR207 pKa = 11.84KK208 pKa = 8.81RR209 pKa = 11.84LYY211 pKa = 10.36CKK213 pKa = 10.32QIQQDD218 pKa = 3.54TDD220 pKa = 3.83TQWFDD225 pKa = 3.66GYY227 pKa = 11.02DD228 pKa = 3.53GQEE231 pKa = 3.8ILLLDD236 pKa = 3.92DD237 pKa = 5.19FGRR240 pKa = 11.84KK241 pKa = 8.7QIGYY245 pKa = 8.59RR246 pKa = 11.84QLLTYY251 pKa = 10.66LDD253 pKa = 3.85IYY255 pKa = 10.18ALKK258 pKa = 10.56VQIKK262 pKa = 10.16GDD264 pKa = 3.69TVDD267 pKa = 4.28ARR269 pKa = 11.84WNYY272 pKa = 10.22VIITTNEE279 pKa = 4.31EE280 pKa = 3.64IQEE283 pKa = 4.14WYY285 pKa = 9.46PNEE288 pKa = 5.25DD289 pKa = 2.83IAPLRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84IAKK299 pKa = 9.03IHH301 pKa = 5.52EE302 pKa = 4.68CNRR305 pKa = 11.84PWAVDD310 pKa = 3.36YY311 pKa = 11.76VDD313 pKa = 5.28FKK315 pKa = 11.41RR316 pKa = 11.84PLPVPVVPQFVLPPEE331 pKa = 4.5PEE333 pKa = 3.88GKK335 pKa = 10.35YY336 pKa = 9.37PQPEE340 pKa = 4.3PNQQAHH346 pKa = 6.63AVPIVDD352 pKa = 3.37EE353 pKa = 4.45VEE355 pKa = 4.36YY356 pKa = 10.83EE357 pKa = 4.06DD358 pKa = 4.72LVPVEE363 pKa = 5.59DD364 pKa = 4.99LDD366 pKa = 5.33ADD368 pKa = 4.01ALIDD372 pKa = 4.63EE373 pKa = 5.05INAEE377 pKa = 4.25LGAIEE382 pKa = 5.21DD383 pKa = 4.11PPDD386 pKa = 3.93FEE388 pKa = 4.65MADD391 pKa = 3.53LGHH394 pKa = 6.66LVWRR398 pKa = 11.84PMM400 pKa = 3.63
MM1 pKa = 7.8PNGSKK6 pKa = 10.39RR7 pKa = 11.84FCFTHH12 pKa = 6.69FGCPDD17 pKa = 3.45EE18 pKa = 4.47FTNILQARR26 pKa = 11.84SPLCTKK32 pKa = 10.02WIVQQEE38 pKa = 4.05RR39 pKa = 11.84GQAEE43 pKa = 4.18GKK45 pKa = 10.69APLEE49 pKa = 4.28GANAQNGGLHH59 pKa = 5.58LQGYY63 pKa = 8.22IHH65 pKa = 6.8TKK67 pKa = 9.58RR68 pKa = 11.84VYY70 pKa = 10.13TFAQFLRR77 pKa = 11.84IWPVSHH83 pKa = 7.09VEE85 pKa = 3.79AAQGTEE91 pKa = 3.73AQNYY95 pKa = 7.44LYY97 pKa = 10.39CRR99 pKa = 11.84KK100 pKa = 10.2NEE102 pKa = 4.13TFTGKK107 pKa = 10.14YY108 pKa = 8.15RR109 pKa = 11.84LAMEE113 pKa = 4.65GSVWSRR119 pKa = 11.84DD120 pKa = 3.75VQRR123 pKa = 11.84LQAKK127 pKa = 8.34SRR129 pKa = 11.84KK130 pKa = 8.39DD131 pKa = 3.46AEE133 pKa = 4.06MQAIVDD139 pKa = 4.45KK140 pKa = 10.39IKK142 pKa = 10.88QGATTADD149 pKa = 3.02IARR152 pKa = 11.84NNTAYY157 pKa = 10.58FIRR160 pKa = 11.84HH161 pKa = 6.12CNGIPAAVRR170 pKa = 11.84HH171 pKa = 5.81HH172 pKa = 6.3QPAAPATRR180 pKa = 11.84PVHH183 pKa = 6.42VILLKK188 pKa = 10.93GSTGTGKK195 pKa = 10.6SYY197 pKa = 9.83WARR200 pKa = 11.84QLATYY205 pKa = 7.99QRR207 pKa = 11.84KK208 pKa = 8.81RR209 pKa = 11.84LYY211 pKa = 10.36CKK213 pKa = 10.32QIQQDD218 pKa = 3.54TDD220 pKa = 3.83TQWFDD225 pKa = 3.66GYY227 pKa = 11.02DD228 pKa = 3.53GQEE231 pKa = 3.8ILLLDD236 pKa = 3.92DD237 pKa = 5.19FGRR240 pKa = 11.84KK241 pKa = 8.7QIGYY245 pKa = 8.59RR246 pKa = 11.84QLLTYY251 pKa = 10.66LDD253 pKa = 3.85IYY255 pKa = 10.18ALKK258 pKa = 10.56VQIKK262 pKa = 10.16GDD264 pKa = 3.69TVDD267 pKa = 4.28ARR269 pKa = 11.84WNYY272 pKa = 10.22VIITTNEE279 pKa = 4.31EE280 pKa = 3.64IQEE283 pKa = 4.14WYY285 pKa = 9.46PNEE288 pKa = 5.25DD289 pKa = 2.83IAPLRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84IAKK299 pKa = 9.03IHH301 pKa = 5.52EE302 pKa = 4.68CNRR305 pKa = 11.84PWAVDD310 pKa = 3.36YY311 pKa = 11.76VDD313 pKa = 5.28FKK315 pKa = 11.41RR316 pKa = 11.84PLPVPVVPQFVLPPEE331 pKa = 4.5PEE333 pKa = 3.88GKK335 pKa = 10.35YY336 pKa = 9.37PQPEE340 pKa = 4.3PNQQAHH346 pKa = 6.63AVPIVDD352 pKa = 3.37EE353 pKa = 4.45VEE355 pKa = 4.36YY356 pKa = 10.83EE357 pKa = 4.06DD358 pKa = 4.72LVPVEE363 pKa = 5.59DD364 pKa = 4.99LDD366 pKa = 5.33ADD368 pKa = 4.01ALIDD372 pKa = 4.63EE373 pKa = 5.05INAEE377 pKa = 4.25LGAIEE382 pKa = 5.21DD383 pKa = 4.11PPDD386 pKa = 3.93FEE388 pKa = 4.65MADD391 pKa = 3.53LGHH394 pKa = 6.66LVWRR398 pKa = 11.84PMM400 pKa = 3.63
Molecular weight: 45.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4YQ33|A0A2H4YQ33_9VIRU Putative capsid protein OS=Idotea virus IWaV278 OX=2058759 PE=3 SV=1
MM1 pKa = 7.12SKK3 pKa = 10.12KK4 pKa = 10.29RR5 pKa = 11.84KK6 pKa = 7.03TVTGGGSRR14 pKa = 11.84RR15 pKa = 11.84STGVPYY21 pKa = 9.86WQMKK25 pKa = 9.4AARR28 pKa = 11.84QYY30 pKa = 11.0RR31 pKa = 11.84DD32 pKa = 3.97DD33 pKa = 4.67YY34 pKa = 9.34MAEE37 pKa = 4.07YY38 pKa = 9.5PRR40 pKa = 11.84GSAASKK46 pKa = 11.05AMWGDD51 pKa = 3.87DD52 pKa = 3.37YY53 pKa = 11.82KK54 pKa = 11.23SASKK58 pKa = 9.95RR59 pKa = 11.84QRR61 pKa = 11.84YY62 pKa = 8.14LRR64 pKa = 11.84NAMRR68 pKa = 11.84FKK70 pKa = 11.33GKK72 pKa = 10.28GKK74 pKa = 10.48YY75 pKa = 8.02SAKK78 pKa = 10.54NFGRR82 pKa = 11.84DD83 pKa = 3.0ALRR86 pKa = 11.84YY87 pKa = 9.76GGAALGGGIGYY98 pKa = 7.44MTGGLAGAGAMAQRR112 pKa = 11.84GYY114 pKa = 9.8DD115 pKa = 3.22TGADD119 pKa = 3.35LSRR122 pKa = 11.84FVGFGEE128 pKa = 4.16YY129 pKa = 9.94KK130 pKa = 10.55GANQLIAGPEE140 pKa = 3.9QNMSITVNRR149 pKa = 11.84EE150 pKa = 3.44HH151 pKa = 7.58RR152 pKa = 11.84SGDD155 pKa = 3.01IVFANTEE162 pKa = 4.11FVQNVYY168 pKa = 10.97AGVTSGPSTSKK179 pKa = 10.55FQTQAFSLNPGLGEE193 pKa = 4.06TFPFLSQIAQNFEE206 pKa = 4.02LFEE209 pKa = 4.27FEE211 pKa = 5.18GLMFEE216 pKa = 4.55YY217 pKa = 10.41KK218 pKa = 8.85PTSGEE223 pKa = 4.01FGTTSSNSLGKK234 pKa = 10.44VVMCTNYY241 pKa = 10.59DD242 pKa = 3.39PEE244 pKa = 5.68APEE247 pKa = 4.19FQNSVQMEE255 pKa = 4.32NYY257 pKa = 10.33DD258 pKa = 3.73YY259 pKa = 11.84ANSTKK264 pKa = 9.92PSRR267 pKa = 11.84GAIHH271 pKa = 6.63GVEE274 pKa = 3.97THH276 pKa = 6.87PDD278 pKa = 3.01QRR280 pKa = 11.84ATKK283 pKa = 10.14LMYY286 pKa = 10.62VRR288 pKa = 11.84TGDD291 pKa = 3.57ANKK294 pKa = 10.75SKK296 pKa = 10.86LFTDD300 pKa = 4.46LGTLHH305 pKa = 7.55LATEE309 pKa = 5.05GVPFGGDD316 pKa = 3.31GQQKK320 pKa = 10.75ALIGEE325 pKa = 4.29LWVTYY330 pKa = 9.81KK331 pKa = 10.57VRR333 pKa = 11.84LSRR336 pKa = 11.84SQLYY340 pKa = 10.83SSVLGNNVITSYY352 pKa = 11.28QSGTSDD358 pKa = 3.15SGVWTGTKK366 pKa = 10.04QFDD369 pKa = 4.03SNTWDD374 pKa = 3.05MSLTNVSQGVFKK386 pKa = 9.86FTLPEE391 pKa = 4.27DD392 pKa = 3.84VDD394 pKa = 4.29DD395 pKa = 5.25GSFLVTISNSSTGTHH410 pKa = 5.17TMSPLEE416 pKa = 4.04FTLEE420 pKa = 3.83NCKK423 pKa = 8.69FTKK426 pKa = 10.0TPVAIAPPPSQVQGWNRR443 pKa = 11.84TYY445 pKa = 11.36LMKK448 pKa = 10.45VDD450 pKa = 3.99AQSALNICSFQFDD463 pKa = 4.71DD464 pKa = 5.77PDD466 pKa = 4.75SNWPNGSWQVGITQVSDD483 pKa = 3.0KK484 pKa = 11.28LYY486 pKa = 10.83DD487 pKa = 3.1EE488 pKa = 4.48WAAFSGEE495 pKa = 4.04LPAA498 pKa = 6.55
MM1 pKa = 7.12SKK3 pKa = 10.12KK4 pKa = 10.29RR5 pKa = 11.84KK6 pKa = 7.03TVTGGGSRR14 pKa = 11.84RR15 pKa = 11.84STGVPYY21 pKa = 9.86WQMKK25 pKa = 9.4AARR28 pKa = 11.84QYY30 pKa = 11.0RR31 pKa = 11.84DD32 pKa = 3.97DD33 pKa = 4.67YY34 pKa = 9.34MAEE37 pKa = 4.07YY38 pKa = 9.5PRR40 pKa = 11.84GSAASKK46 pKa = 11.05AMWGDD51 pKa = 3.87DD52 pKa = 3.37YY53 pKa = 11.82KK54 pKa = 11.23SASKK58 pKa = 9.95RR59 pKa = 11.84QRR61 pKa = 11.84YY62 pKa = 8.14LRR64 pKa = 11.84NAMRR68 pKa = 11.84FKK70 pKa = 11.33GKK72 pKa = 10.28GKK74 pKa = 10.48YY75 pKa = 8.02SAKK78 pKa = 10.54NFGRR82 pKa = 11.84DD83 pKa = 3.0ALRR86 pKa = 11.84YY87 pKa = 9.76GGAALGGGIGYY98 pKa = 7.44MTGGLAGAGAMAQRR112 pKa = 11.84GYY114 pKa = 9.8DD115 pKa = 3.22TGADD119 pKa = 3.35LSRR122 pKa = 11.84FVGFGEE128 pKa = 4.16YY129 pKa = 9.94KK130 pKa = 10.55GANQLIAGPEE140 pKa = 3.9QNMSITVNRR149 pKa = 11.84EE150 pKa = 3.44HH151 pKa = 7.58RR152 pKa = 11.84SGDD155 pKa = 3.01IVFANTEE162 pKa = 4.11FVQNVYY168 pKa = 10.97AGVTSGPSTSKK179 pKa = 10.55FQTQAFSLNPGLGEE193 pKa = 4.06TFPFLSQIAQNFEE206 pKa = 4.02LFEE209 pKa = 4.27FEE211 pKa = 5.18GLMFEE216 pKa = 4.55YY217 pKa = 10.41KK218 pKa = 8.85PTSGEE223 pKa = 4.01FGTTSSNSLGKK234 pKa = 10.44VVMCTNYY241 pKa = 10.59DD242 pKa = 3.39PEE244 pKa = 5.68APEE247 pKa = 4.19FQNSVQMEE255 pKa = 4.32NYY257 pKa = 10.33DD258 pKa = 3.73YY259 pKa = 11.84ANSTKK264 pKa = 9.92PSRR267 pKa = 11.84GAIHH271 pKa = 6.63GVEE274 pKa = 3.97THH276 pKa = 6.87PDD278 pKa = 3.01QRR280 pKa = 11.84ATKK283 pKa = 10.14LMYY286 pKa = 10.62VRR288 pKa = 11.84TGDD291 pKa = 3.57ANKK294 pKa = 10.75SKK296 pKa = 10.86LFTDD300 pKa = 4.46LGTLHH305 pKa = 7.55LATEE309 pKa = 5.05GVPFGGDD316 pKa = 3.31GQQKK320 pKa = 10.75ALIGEE325 pKa = 4.29LWVTYY330 pKa = 9.81KK331 pKa = 10.57VRR333 pKa = 11.84LSRR336 pKa = 11.84SQLYY340 pKa = 10.83SSVLGNNVITSYY352 pKa = 11.28QSGTSDD358 pKa = 3.15SGVWTGTKK366 pKa = 10.04QFDD369 pKa = 4.03SNTWDD374 pKa = 3.05MSLTNVSQGVFKK386 pKa = 9.86FTLPEE391 pKa = 4.27DD392 pKa = 3.84VDD394 pKa = 4.29DD395 pKa = 5.25GSFLVTISNSSTGTHH410 pKa = 5.17TMSPLEE416 pKa = 4.04FTLEE420 pKa = 3.83NCKK423 pKa = 8.69FTKK426 pKa = 10.0TPVAIAPPPSQVQGWNRR443 pKa = 11.84TYY445 pKa = 11.36LMKK448 pKa = 10.45VDD450 pKa = 3.99AQSALNICSFQFDD463 pKa = 4.71DD464 pKa = 5.77PDD466 pKa = 4.75SNWPNGSWQVGITQVSDD483 pKa = 3.0KK484 pKa = 11.28LYY486 pKa = 10.83DD487 pKa = 3.1EE488 pKa = 4.48WAAFSGEE495 pKa = 4.04LPAA498 pKa = 6.55
Molecular weight: 54.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
898 |
400 |
498 |
449.0 |
50.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.129 ± 0.536 | 1.114 ± 0.392 |
5.791 ± 0.437 | 5.345 ± 0.711 |
4.454 ± 0.742 | 8.686 ± 1.654 |
1.782 ± 0.596 | 4.12 ± 1.311 |
5.234 ± 0.144 | 6.682 ± 0.35 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.227 ± 0.602 | 4.454 ± 0.434 |
5.345 ± 0.865 | 6.125 ± 0.693 |
5.345 ± 0.711 | 6.125 ± 2.54 |
6.682 ± 0.881 | 5.902 ± 0.214 |
2.004 ± 0.151 | 4.454 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
