
Bosea thiooxidans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Boseaceae; Bosea
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
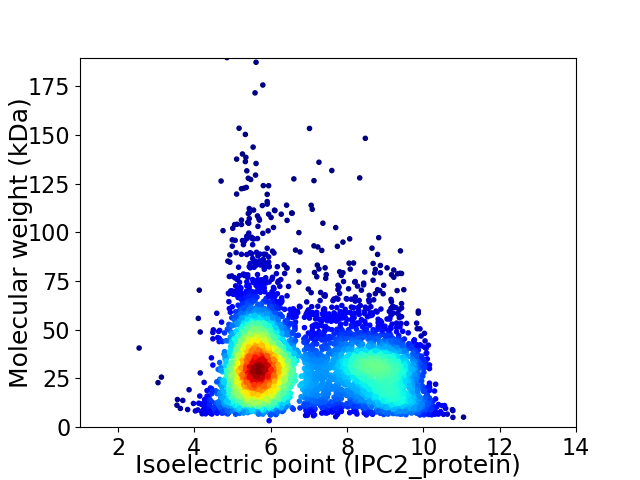
Virtual 2D-PAGE plot for 4956 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q3HZ22|A0A0Q3HZ22_9BRAD Aminopeptidase N OS=Bosea thiooxidans OX=53254 GN=pepN PE=3 SV=1
MM1 pKa = 8.09PDD3 pKa = 2.98LTGYY7 pKa = 11.0DD8 pKa = 3.47LVLASTEE15 pKa = 4.1SALNSSIAYY24 pKa = 9.82ANFWKK29 pKa = 9.07VTINCQLNPDD39 pKa = 3.77VSAEE43 pKa = 3.95QSPGLVGARR52 pKa = 11.84ISAPTLRR59 pKa = 11.84LSGSDD64 pKa = 3.27LASNQAAVSLLFTAGSLQYY83 pKa = 10.65FDD85 pKa = 4.29VIYY88 pKa = 10.84YY89 pKa = 7.88RR90 pKa = 11.84TVSQNIAGWTIVFAANIGQGAIPDD114 pKa = 3.74NTEE117 pKa = 3.49IGQLPAEE124 pKa = 4.4VQNALSNQRR133 pKa = 11.84LSVGQLFLEE142 pKa = 4.88LSTVVWANVTITDD155 pKa = 3.72ADD157 pKa = 3.92GKK159 pKa = 10.48RR160 pKa = 11.84VTDD163 pKa = 5.09AIITSTLEE171 pKa = 3.61TALQSWARR179 pKa = 11.84SIEE182 pKa = 4.14SAAQQWVLNYY192 pKa = 10.07VVTSSGGPGNASGEE206 pKa = 4.31VLADD210 pKa = 4.16LVPTSYY216 pKa = 11.27DD217 pKa = 3.42FSVSKK222 pKa = 10.47YY223 pKa = 9.74NPPGTYY229 pKa = 10.3DD230 pKa = 4.53DD231 pKa = 5.82LSTLNYY237 pKa = 11.12LMMTEE242 pKa = 4.35GRR244 pKa = 11.84GAPTANSAGIFDD256 pKa = 5.02FNWVAGPNTQQYY268 pKa = 9.5QGAVAINGPLFDD280 pKa = 3.92GAYY283 pKa = 10.04VEE285 pKa = 4.77PLLLPAIAAAVSATKK300 pKa = 8.73PTSAGGSTGNPGVPTFTRR318 pKa = 11.84TGPGQWSTSQSVDD331 pKa = 3.32LYY333 pKa = 11.16GNQHH337 pKa = 5.82GGKK340 pKa = 8.57GQKK343 pKa = 10.31LEE345 pKa = 4.09GLGLLQAVYY354 pKa = 10.99QNITEE359 pKa = 4.42KK360 pKa = 10.88LDD362 pKa = 3.54CTVTLDD368 pKa = 4.02PASATYY374 pKa = 8.94QVKK377 pKa = 8.09VTYY380 pKa = 10.37SAITTITVEE389 pKa = 3.76FFGWDD394 pKa = 3.14KK395 pKa = 10.94TDD397 pKa = 3.51SSFSGDD403 pKa = 3.89YY404 pKa = 10.17VLPWSFDD411 pKa = 3.41VRR413 pKa = 11.84LKK415 pKa = 10.91ASSNGSLVTEE425 pKa = 4.12STEE428 pKa = 4.05VQEE431 pKa = 5.15GEE433 pKa = 4.51SSSKK437 pKa = 8.38TWGNSFVGDD446 pKa = 3.14IWATISEE453 pKa = 4.51VFSSFLNTYY462 pKa = 7.41QTQFQDD468 pKa = 3.1KK469 pKa = 9.2AASVSQSAFANVSDD483 pKa = 4.14EE484 pKa = 4.77LEE486 pKa = 4.12SAMGDD491 pKa = 2.93ISAAIVFPAGNRR503 pKa = 11.84FAYY506 pKa = 9.8KK507 pKa = 10.06DD508 pKa = 3.46VCFDD512 pKa = 3.56PQQNLVAYY520 pKa = 7.46VTYY523 pKa = 11.04LNN525 pKa = 4.08
MM1 pKa = 8.09PDD3 pKa = 2.98LTGYY7 pKa = 11.0DD8 pKa = 3.47LVLASTEE15 pKa = 4.1SALNSSIAYY24 pKa = 9.82ANFWKK29 pKa = 9.07VTINCQLNPDD39 pKa = 3.77VSAEE43 pKa = 3.95QSPGLVGARR52 pKa = 11.84ISAPTLRR59 pKa = 11.84LSGSDD64 pKa = 3.27LASNQAAVSLLFTAGSLQYY83 pKa = 10.65FDD85 pKa = 4.29VIYY88 pKa = 10.84YY89 pKa = 7.88RR90 pKa = 11.84TVSQNIAGWTIVFAANIGQGAIPDD114 pKa = 3.74NTEE117 pKa = 3.49IGQLPAEE124 pKa = 4.4VQNALSNQRR133 pKa = 11.84LSVGQLFLEE142 pKa = 4.88LSTVVWANVTITDD155 pKa = 3.72ADD157 pKa = 3.92GKK159 pKa = 10.48RR160 pKa = 11.84VTDD163 pKa = 5.09AIITSTLEE171 pKa = 3.61TALQSWARR179 pKa = 11.84SIEE182 pKa = 4.14SAAQQWVLNYY192 pKa = 10.07VVTSSGGPGNASGEE206 pKa = 4.31VLADD210 pKa = 4.16LVPTSYY216 pKa = 11.27DD217 pKa = 3.42FSVSKK222 pKa = 10.47YY223 pKa = 9.74NPPGTYY229 pKa = 10.3DD230 pKa = 4.53DD231 pKa = 5.82LSTLNYY237 pKa = 11.12LMMTEE242 pKa = 4.35GRR244 pKa = 11.84GAPTANSAGIFDD256 pKa = 5.02FNWVAGPNTQQYY268 pKa = 9.5QGAVAINGPLFDD280 pKa = 3.92GAYY283 pKa = 10.04VEE285 pKa = 4.77PLLLPAIAAAVSATKK300 pKa = 8.73PTSAGGSTGNPGVPTFTRR318 pKa = 11.84TGPGQWSTSQSVDD331 pKa = 3.32LYY333 pKa = 11.16GNQHH337 pKa = 5.82GGKK340 pKa = 8.57GQKK343 pKa = 10.31LEE345 pKa = 4.09GLGLLQAVYY354 pKa = 10.99QNITEE359 pKa = 4.42KK360 pKa = 10.88LDD362 pKa = 3.54CTVTLDD368 pKa = 4.02PASATYY374 pKa = 8.94QVKK377 pKa = 8.09VTYY380 pKa = 10.37SAITTITVEE389 pKa = 3.76FFGWDD394 pKa = 3.14KK395 pKa = 10.94TDD397 pKa = 3.51SSFSGDD403 pKa = 3.89YY404 pKa = 10.17VLPWSFDD411 pKa = 3.41VRR413 pKa = 11.84LKK415 pKa = 10.91ASSNGSLVTEE425 pKa = 4.12STEE428 pKa = 4.05VQEE431 pKa = 5.15GEE433 pKa = 4.51SSSKK437 pKa = 8.38TWGNSFVGDD446 pKa = 3.14IWATISEE453 pKa = 4.51VFSSFLNTYY462 pKa = 7.41QTQFQDD468 pKa = 3.1KK469 pKa = 9.2AASVSQSAFANVSDD483 pKa = 4.14EE484 pKa = 4.77LEE486 pKa = 4.12SAMGDD491 pKa = 2.93ISAAIVFPAGNRR503 pKa = 11.84FAYY506 pKa = 9.8KK507 pKa = 10.06DD508 pKa = 3.46VCFDD512 pKa = 3.56PQQNLVAYY520 pKa = 7.46VTYY523 pKa = 11.04LNN525 pKa = 4.08
Molecular weight: 55.84 kDa
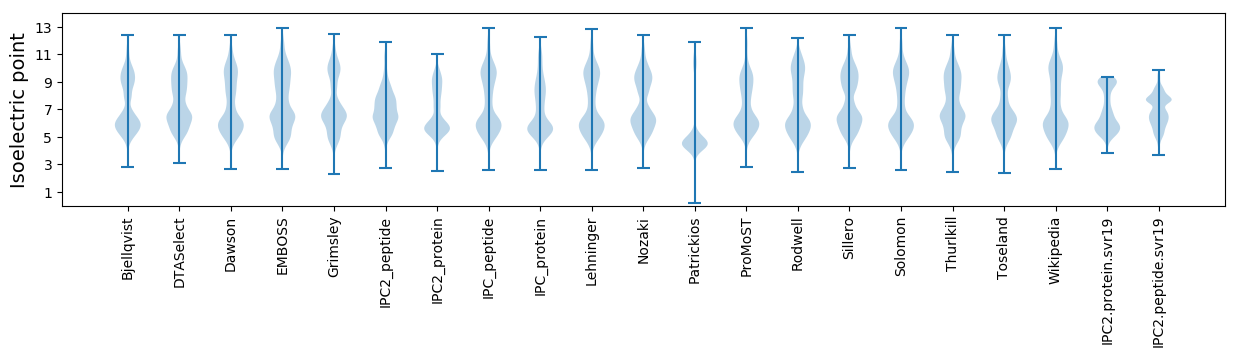
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q3M325|A0A0Q3M325_9BRAD DNA-binding transcriptional regulator GntR family OS=Bosea thiooxidans OX=53254 GN=ARD30_14750 PE=4 SV=1
MM1 pKa = 7.6SARR4 pKa = 11.84LPSLLLLVALLAGWEE19 pKa = 4.12AWCRR23 pKa = 11.84LGGVSALVLPPPSAVLATLWSEE45 pKa = 3.94IATGRR50 pKa = 11.84LWPHH54 pKa = 6.58LAVTATEE61 pKa = 4.15MALGLAIGAVVGLGVGILLAEE82 pKa = 4.88FPALSRR88 pKa = 11.84LLRR91 pKa = 11.84PYY93 pKa = 10.69VLASQLVPKK102 pKa = 10.21LALGPLFIIWFGFGMTPTVVITALICFFPLMEE134 pKa = 4.27NTLTGVSEE142 pKa = 3.98VDD144 pKa = 3.21PARR147 pKa = 11.84RR148 pKa = 11.84EE149 pKa = 4.02LFRR152 pKa = 11.84MLGASRR158 pKa = 11.84LQTLLRR164 pKa = 11.84LKK166 pKa = 10.63LPAALPVIMAGLRR179 pKa = 11.84VAVVLALVGAVVGEE193 pKa = 4.46FIGGRR198 pKa = 11.84IGLGASIIAAQSVMDD213 pKa = 4.03SSLIFALFIVITALGMLLYY232 pKa = 10.34EE233 pKa = 4.4AVRR236 pKa = 11.84LLEE239 pKa = 3.82RR240 pKa = 11.84RR241 pKa = 11.84VLHH244 pKa = 6.35RR245 pKa = 11.84FSKK248 pKa = 10.8GG249 pKa = 3.0
MM1 pKa = 7.6SARR4 pKa = 11.84LPSLLLLVALLAGWEE19 pKa = 4.12AWCRR23 pKa = 11.84LGGVSALVLPPPSAVLATLWSEE45 pKa = 3.94IATGRR50 pKa = 11.84LWPHH54 pKa = 6.58LAVTATEE61 pKa = 4.15MALGLAIGAVVGLGVGILLAEE82 pKa = 4.88FPALSRR88 pKa = 11.84LLRR91 pKa = 11.84PYY93 pKa = 10.69VLASQLVPKK102 pKa = 10.21LALGPLFIIWFGFGMTPTVVITALICFFPLMEE134 pKa = 4.27NTLTGVSEE142 pKa = 3.98VDD144 pKa = 3.21PARR147 pKa = 11.84RR148 pKa = 11.84EE149 pKa = 4.02LFRR152 pKa = 11.84MLGASRR158 pKa = 11.84LQTLLRR164 pKa = 11.84LKK166 pKa = 10.63LPAALPVIMAGLRR179 pKa = 11.84VAVVLALVGAVVGEE193 pKa = 4.46FIGGRR198 pKa = 11.84IGLGASIIAAQSVMDD213 pKa = 4.03SSLIFALFIVITALGMLLYY232 pKa = 10.34EE233 pKa = 4.4AVRR236 pKa = 11.84LLEE239 pKa = 3.82RR240 pKa = 11.84RR241 pKa = 11.84VLHH244 pKa = 6.35RR245 pKa = 11.84FSKK248 pKa = 10.8GG249 pKa = 3.0
Molecular weight: 26.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1551163 |
29 |
1793 |
313.0 |
33.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.603 ± 0.044 | 0.797 ± 0.009 |
5.23 ± 0.028 | 5.593 ± 0.03 |
3.708 ± 0.021 | 8.903 ± 0.035 |
1.918 ± 0.017 | 5.108 ± 0.024 |
3.16 ± 0.029 | 10.641 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.364 ± 0.015 | 2.286 ± 0.017 |
5.39 ± 0.027 | 3.013 ± 0.021 |
7.335 ± 0.036 | 5.099 ± 0.022 |
5.003 ± 0.021 | 7.446 ± 0.028 |
1.328 ± 0.013 | 2.074 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
