
Muir Springs virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Barhavirus; Muir barhavirus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
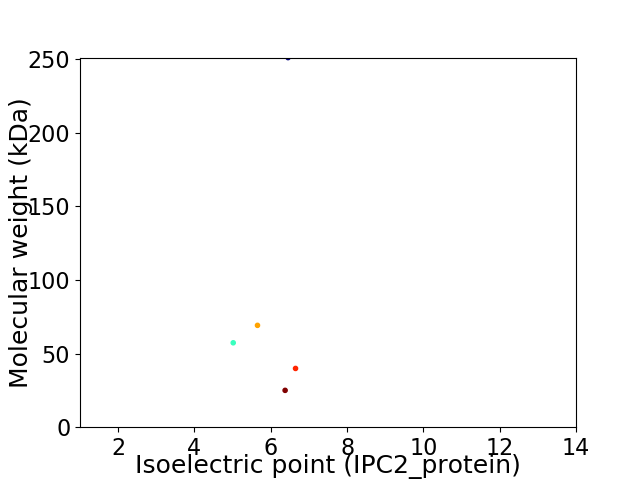
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3R160|A0A0D3R160_9RHAB GDP polyribonucleotidyltransferase OS=Muir Springs virus OX=932700 PE=4 SV=1
MM1 pKa = 7.49AVAILPVSRR10 pKa = 11.84NLPARR15 pKa = 11.84NRR17 pKa = 11.84TVAGSVTAPPVQYY30 pKa = 9.72PSEE33 pKa = 3.9WFRR36 pKa = 11.84ANEE39 pKa = 3.76NRR41 pKa = 11.84KK42 pKa = 9.64VSITIYY48 pKa = 10.01QQTTAGQAYY57 pKa = 10.16SRR59 pKa = 11.84IEE61 pKa = 3.64ALRR64 pKa = 11.84NNGQWDD70 pKa = 4.0DD71 pKa = 3.86VLISTFMRR79 pKa = 11.84GVIDD83 pKa = 5.48EE84 pKa = 4.15NAEE87 pKa = 4.04WFQSAPLTEE96 pKa = 4.35DD97 pKa = 2.66WSVNGAVIGQTGASITPLNLATWEE121 pKa = 4.27TIPRR125 pKa = 11.84PGNLGPIANQEE136 pKa = 4.19GEE138 pKa = 4.31LEE140 pKa = 4.18TRR142 pKa = 11.84RR143 pKa = 11.84SFFLALVTIYY153 pKa = 10.55RR154 pKa = 11.84QVLTRR159 pKa = 11.84DD160 pKa = 2.91INAEE164 pKa = 3.83YY165 pKa = 9.52GQEE168 pKa = 3.75VSRR171 pKa = 11.84RR172 pKa = 11.84IIDD175 pKa = 3.59NFKK178 pKa = 9.86EE179 pKa = 3.98QPLAMSQDD187 pKa = 4.5DD188 pKa = 3.95INNIQNFEE196 pKa = 4.3TKK198 pKa = 10.46DD199 pKa = 3.56KK200 pKa = 10.45LTANYY205 pKa = 10.21VKK207 pKa = 10.28VLCILDD213 pKa = 3.32MFFNKK218 pKa = 9.44FQNHH222 pKa = 6.72DD223 pKa = 3.27KK224 pKa = 9.88STIRR228 pKa = 11.84IATLPTRR235 pKa = 11.84YY236 pKa = 9.38RR237 pKa = 11.84GCAAFTSYY245 pKa = 11.65GEE247 pKa = 3.81LAIRR251 pKa = 11.84LGIEE255 pKa = 4.05PVQLPSLILTMAVARR270 pKa = 11.84DD271 pKa = 3.47FDD273 pKa = 4.49KK274 pKa = 11.26INIGGEE280 pKa = 4.09QAEE283 pKa = 4.42HH284 pKa = 6.8LDD286 pKa = 4.08GYY288 pKa = 10.88FPYY291 pKa = 10.62QLTLGLVKK299 pKa = 10.4KK300 pKa = 10.24SAYY303 pKa = 9.7SASNCPALYY312 pKa = 10.41LWMHH316 pKa = 7.01TIGTMLYY323 pKa = 7.42QQRR326 pKa = 11.84SYY328 pKa = 10.8RR329 pKa = 11.84AQVPKK334 pKa = 10.69NVPDD338 pKa = 3.36QMGTINSAIAVAIQYY353 pKa = 9.04VAGGDD358 pKa = 3.59FSMQFVADD366 pKa = 3.81ARR368 pKa = 11.84VQGAMRR374 pKa = 11.84EE375 pKa = 4.24MQDD378 pKa = 3.12AEE380 pKa = 4.55AEE382 pKa = 4.34LTHH385 pKa = 6.73LRR387 pKa = 11.84QLTARR392 pKa = 11.84AARR395 pKa = 11.84QAAEE399 pKa = 4.2GNVNDD404 pKa = 4.04EE405 pKa = 4.72GGDD408 pKa = 3.52EE409 pKa = 4.54GEE411 pKa = 4.2SSEE414 pKa = 5.57DD415 pKa = 3.39EE416 pKa = 4.37EE417 pKa = 5.89EE418 pKa = 4.49EE419 pKa = 5.63DD420 pKa = 5.13DD421 pKa = 5.43HH422 pKa = 8.95QLDD425 pKa = 4.39DD426 pKa = 4.6QDD428 pKa = 4.68DD429 pKa = 4.13PEE431 pKa = 4.6IAARR435 pKa = 11.84LEE437 pKa = 4.48RR438 pKa = 11.84IGVLQEE444 pKa = 3.41NLRR447 pKa = 11.84RR448 pKa = 11.84CTATVEE454 pKa = 4.22QFTSVVEE461 pKa = 4.21KK462 pKa = 10.78SAMRR466 pKa = 11.84ALAYY470 pKa = 10.14LQEE473 pKa = 4.43NGGIAEE479 pKa = 4.14KK480 pKa = 10.57DD481 pKa = 3.25RR482 pKa = 11.84RR483 pKa = 11.84EE484 pKa = 3.67LGIRR488 pKa = 11.84FRR490 pKa = 11.84RR491 pKa = 11.84FADD494 pKa = 3.61EE495 pKa = 3.82ADD497 pKa = 3.45GRR499 pKa = 11.84VGKK502 pKa = 10.29LLANLFPAHH511 pKa = 6.77RR512 pKa = 4.62
MM1 pKa = 7.49AVAILPVSRR10 pKa = 11.84NLPARR15 pKa = 11.84NRR17 pKa = 11.84TVAGSVTAPPVQYY30 pKa = 9.72PSEE33 pKa = 3.9WFRR36 pKa = 11.84ANEE39 pKa = 3.76NRR41 pKa = 11.84KK42 pKa = 9.64VSITIYY48 pKa = 10.01QQTTAGQAYY57 pKa = 10.16SRR59 pKa = 11.84IEE61 pKa = 3.64ALRR64 pKa = 11.84NNGQWDD70 pKa = 4.0DD71 pKa = 3.86VLISTFMRR79 pKa = 11.84GVIDD83 pKa = 5.48EE84 pKa = 4.15NAEE87 pKa = 4.04WFQSAPLTEE96 pKa = 4.35DD97 pKa = 2.66WSVNGAVIGQTGASITPLNLATWEE121 pKa = 4.27TIPRR125 pKa = 11.84PGNLGPIANQEE136 pKa = 4.19GEE138 pKa = 4.31LEE140 pKa = 4.18TRR142 pKa = 11.84RR143 pKa = 11.84SFFLALVTIYY153 pKa = 10.55RR154 pKa = 11.84QVLTRR159 pKa = 11.84DD160 pKa = 2.91INAEE164 pKa = 3.83YY165 pKa = 9.52GQEE168 pKa = 3.75VSRR171 pKa = 11.84RR172 pKa = 11.84IIDD175 pKa = 3.59NFKK178 pKa = 9.86EE179 pKa = 3.98QPLAMSQDD187 pKa = 4.5DD188 pKa = 3.95INNIQNFEE196 pKa = 4.3TKK198 pKa = 10.46DD199 pKa = 3.56KK200 pKa = 10.45LTANYY205 pKa = 10.21VKK207 pKa = 10.28VLCILDD213 pKa = 3.32MFFNKK218 pKa = 9.44FQNHH222 pKa = 6.72DD223 pKa = 3.27KK224 pKa = 9.88STIRR228 pKa = 11.84IATLPTRR235 pKa = 11.84YY236 pKa = 9.38RR237 pKa = 11.84GCAAFTSYY245 pKa = 11.65GEE247 pKa = 3.81LAIRR251 pKa = 11.84LGIEE255 pKa = 4.05PVQLPSLILTMAVARR270 pKa = 11.84DD271 pKa = 3.47FDD273 pKa = 4.49KK274 pKa = 11.26INIGGEE280 pKa = 4.09QAEE283 pKa = 4.42HH284 pKa = 6.8LDD286 pKa = 4.08GYY288 pKa = 10.88FPYY291 pKa = 10.62QLTLGLVKK299 pKa = 10.4KK300 pKa = 10.24SAYY303 pKa = 9.7SASNCPALYY312 pKa = 10.41LWMHH316 pKa = 7.01TIGTMLYY323 pKa = 7.42QQRR326 pKa = 11.84SYY328 pKa = 10.8RR329 pKa = 11.84AQVPKK334 pKa = 10.69NVPDD338 pKa = 3.36QMGTINSAIAVAIQYY353 pKa = 9.04VAGGDD358 pKa = 3.59FSMQFVADD366 pKa = 3.81ARR368 pKa = 11.84VQGAMRR374 pKa = 11.84EE375 pKa = 4.24MQDD378 pKa = 3.12AEE380 pKa = 4.55AEE382 pKa = 4.34LTHH385 pKa = 6.73LRR387 pKa = 11.84QLTARR392 pKa = 11.84AARR395 pKa = 11.84QAAEE399 pKa = 4.2GNVNDD404 pKa = 4.04EE405 pKa = 4.72GGDD408 pKa = 3.52EE409 pKa = 4.54GEE411 pKa = 4.2SSEE414 pKa = 5.57DD415 pKa = 3.39EE416 pKa = 4.37EE417 pKa = 5.89EE418 pKa = 4.49EE419 pKa = 5.63DD420 pKa = 5.13DD421 pKa = 5.43HH422 pKa = 8.95QLDD425 pKa = 4.39DD426 pKa = 4.6QDD428 pKa = 4.68DD429 pKa = 4.13PEE431 pKa = 4.6IAARR435 pKa = 11.84LEE437 pKa = 4.48RR438 pKa = 11.84IGVLQEE444 pKa = 3.41NLRR447 pKa = 11.84RR448 pKa = 11.84CTATVEE454 pKa = 4.22QFTSVVEE461 pKa = 4.21KK462 pKa = 10.78SAMRR466 pKa = 11.84ALAYY470 pKa = 10.14LQEE473 pKa = 4.43NGGIAEE479 pKa = 4.14KK480 pKa = 10.57DD481 pKa = 3.25RR482 pKa = 11.84RR483 pKa = 11.84EE484 pKa = 3.67LGIRR488 pKa = 11.84FRR490 pKa = 11.84RR491 pKa = 11.84FADD494 pKa = 3.61EE495 pKa = 3.82ADD497 pKa = 3.45GRR499 pKa = 11.84VGKK502 pKa = 10.29LLANLFPAHH511 pKa = 6.77RR512 pKa = 4.62
Molecular weight: 57.31 kDa
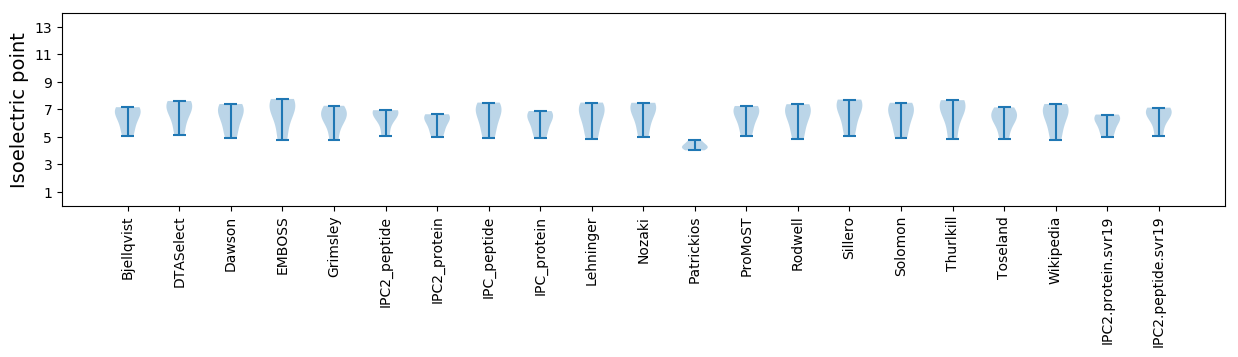
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3R124|A0A0D3R124_9RHAB Nucleocapsid protein OS=Muir Springs virus OX=932700 PE=4 SV=1
MM1 pKa = 7.6SYY3 pKa = 8.9STGLIRR9 pKa = 11.84SEE11 pKa = 4.45VSSEE15 pKa = 3.57MSKK18 pKa = 10.81AFKK21 pKa = 10.43DD22 pKa = 3.53AGEE25 pKa = 4.12HH26 pKa = 5.97QIALNKK32 pKa = 10.38EE33 pKa = 4.03YY34 pKa = 11.03DD35 pKa = 3.66NMSSISTNMNALKK48 pKa = 11.07NMFNPDD54 pKa = 3.94DD55 pKa = 5.07DD56 pKa = 5.22SNSDD60 pKa = 3.3GTSPSSRR67 pKa = 11.84NSILLASDD75 pKa = 3.62MFLGNDD81 pKa = 4.41DD82 pKa = 4.25YY83 pKa = 12.11GSDD86 pKa = 3.78DD87 pKa = 3.26SHH89 pKa = 7.67NFLSSPSPDD98 pKa = 2.94RR99 pKa = 11.84DD100 pKa = 3.55SSEE103 pKa = 4.18EE104 pKa = 3.78GSKK107 pKa = 9.77IQEE110 pKa = 3.89FRR112 pKa = 11.84FEE114 pKa = 4.03IPKK117 pKa = 10.65SKK119 pKa = 9.47TYY121 pKa = 10.63KK122 pKa = 9.75DD123 pKa = 2.83RR124 pKa = 11.84SYY126 pKa = 11.04RR127 pKa = 11.84RR128 pKa = 11.84GVIDD132 pKa = 4.82VLDD135 pKa = 3.95FLQRR139 pKa = 11.84HH140 pKa = 5.36HH141 pKa = 7.0FIEE144 pKa = 4.48EE145 pKa = 3.89FLFEE149 pKa = 4.83GDD151 pKa = 3.42HH152 pKa = 6.43EE153 pKa = 4.89EE154 pKa = 4.99IIVITPTKK162 pKa = 10.75GMIPFKK168 pKa = 10.31RR169 pKa = 11.84SKK171 pKa = 10.98SEE173 pKa = 3.92DD174 pKa = 3.33SQIPMANEE182 pKa = 3.26KK183 pKa = 10.62DD184 pKa = 3.41IEE186 pKa = 4.21KK187 pKa = 10.98EE188 pKa = 4.02EE189 pKa = 3.88IEE191 pKa = 4.7KK192 pKa = 10.25PVKK195 pKa = 10.26SDD197 pKa = 2.99ARR199 pKa = 11.84NTTSEE204 pKa = 4.2HH205 pKa = 5.3STSIEE210 pKa = 3.96DD211 pKa = 3.44PKK213 pKa = 11.45NRR215 pKa = 11.84LKK217 pKa = 10.8EE218 pKa = 3.99ALRR221 pKa = 11.84KK222 pKa = 9.57NIKK225 pKa = 10.16DD226 pKa = 3.6DD227 pKa = 3.84LRR229 pKa = 11.84SKK231 pKa = 10.28ISKK234 pKa = 9.59NVPKK238 pKa = 10.43KK239 pKa = 10.79QEE241 pKa = 3.82TADD244 pKa = 3.79DD245 pKa = 3.88NKK247 pKa = 11.08EE248 pKa = 3.95GVRR251 pKa = 11.84PTNQKK256 pKa = 10.4EE257 pKa = 4.25MIPSQDD263 pKa = 3.1MTKK266 pKa = 8.95STKK269 pKa = 10.05KK270 pKa = 10.17QEE272 pKa = 4.26LSAKK276 pKa = 10.23PPIKK280 pKa = 10.25DD281 pKa = 3.42GKK283 pKa = 10.54KK284 pKa = 10.04KK285 pKa = 9.9EE286 pKa = 4.16DD287 pKa = 3.4KK288 pKa = 10.52KK289 pKa = 11.39AEE291 pKa = 4.08DD292 pKa = 3.26THH294 pKa = 9.47KK295 pKa = 11.0SFNYY299 pKa = 10.5ANTFGSKK306 pKa = 7.62VTVKK310 pKa = 9.82TSKK313 pKa = 10.13YY314 pKa = 10.26CKK316 pKa = 8.76NCKK319 pKa = 9.7AATRR323 pKa = 11.84KK324 pKa = 9.63NATIYY329 pKa = 10.93LNHH332 pKa = 7.47LYY334 pKa = 9.8TNHH337 pKa = 5.37SHH339 pKa = 7.09EE340 pKa = 4.09IALIKK345 pKa = 10.6SLAYY349 pKa = 9.56PLL351 pKa = 4.17
MM1 pKa = 7.6SYY3 pKa = 8.9STGLIRR9 pKa = 11.84SEE11 pKa = 4.45VSSEE15 pKa = 3.57MSKK18 pKa = 10.81AFKK21 pKa = 10.43DD22 pKa = 3.53AGEE25 pKa = 4.12HH26 pKa = 5.97QIALNKK32 pKa = 10.38EE33 pKa = 4.03YY34 pKa = 11.03DD35 pKa = 3.66NMSSISTNMNALKK48 pKa = 11.07NMFNPDD54 pKa = 3.94DD55 pKa = 5.07DD56 pKa = 5.22SNSDD60 pKa = 3.3GTSPSSRR67 pKa = 11.84NSILLASDD75 pKa = 3.62MFLGNDD81 pKa = 4.41DD82 pKa = 4.25YY83 pKa = 12.11GSDD86 pKa = 3.78DD87 pKa = 3.26SHH89 pKa = 7.67NFLSSPSPDD98 pKa = 2.94RR99 pKa = 11.84DD100 pKa = 3.55SSEE103 pKa = 4.18EE104 pKa = 3.78GSKK107 pKa = 9.77IQEE110 pKa = 3.89FRR112 pKa = 11.84FEE114 pKa = 4.03IPKK117 pKa = 10.65SKK119 pKa = 9.47TYY121 pKa = 10.63KK122 pKa = 9.75DD123 pKa = 2.83RR124 pKa = 11.84SYY126 pKa = 11.04RR127 pKa = 11.84RR128 pKa = 11.84GVIDD132 pKa = 4.82VLDD135 pKa = 3.95FLQRR139 pKa = 11.84HH140 pKa = 5.36HH141 pKa = 7.0FIEE144 pKa = 4.48EE145 pKa = 3.89FLFEE149 pKa = 4.83GDD151 pKa = 3.42HH152 pKa = 6.43EE153 pKa = 4.89EE154 pKa = 4.99IIVITPTKK162 pKa = 10.75GMIPFKK168 pKa = 10.31RR169 pKa = 11.84SKK171 pKa = 10.98SEE173 pKa = 3.92DD174 pKa = 3.33SQIPMANEE182 pKa = 3.26KK183 pKa = 10.62DD184 pKa = 3.41IEE186 pKa = 4.21KK187 pKa = 10.98EE188 pKa = 4.02EE189 pKa = 3.88IEE191 pKa = 4.7KK192 pKa = 10.25PVKK195 pKa = 10.26SDD197 pKa = 2.99ARR199 pKa = 11.84NTTSEE204 pKa = 4.2HH205 pKa = 5.3STSIEE210 pKa = 3.96DD211 pKa = 3.44PKK213 pKa = 11.45NRR215 pKa = 11.84LKK217 pKa = 10.8EE218 pKa = 3.99ALRR221 pKa = 11.84KK222 pKa = 9.57NIKK225 pKa = 10.16DD226 pKa = 3.6DD227 pKa = 3.84LRR229 pKa = 11.84SKK231 pKa = 10.28ISKK234 pKa = 9.59NVPKK238 pKa = 10.43KK239 pKa = 10.79QEE241 pKa = 3.82TADD244 pKa = 3.79DD245 pKa = 3.88NKK247 pKa = 11.08EE248 pKa = 3.95GVRR251 pKa = 11.84PTNQKK256 pKa = 10.4EE257 pKa = 4.25MIPSQDD263 pKa = 3.1MTKK266 pKa = 8.95STKK269 pKa = 10.05KK270 pKa = 10.17QEE272 pKa = 4.26LSAKK276 pKa = 10.23PPIKK280 pKa = 10.25DD281 pKa = 3.42GKK283 pKa = 10.54KK284 pKa = 10.04KK285 pKa = 9.9EE286 pKa = 4.16DD287 pKa = 3.4KK288 pKa = 10.52KK289 pKa = 11.39AEE291 pKa = 4.08DD292 pKa = 3.26THH294 pKa = 9.47KK295 pKa = 11.0SFNYY299 pKa = 10.5ANTFGSKK306 pKa = 7.62VTVKK310 pKa = 9.82TSKK313 pKa = 10.13YY314 pKa = 10.26CKK316 pKa = 8.76NCKK319 pKa = 9.7AATRR323 pKa = 11.84KK324 pKa = 9.63NATIYY329 pKa = 10.93LNHH332 pKa = 7.47LYY334 pKa = 9.8TNHH337 pKa = 5.37SHH339 pKa = 7.09EE340 pKa = 4.09IALIKK345 pKa = 10.6SLAYY349 pKa = 9.56PLL351 pKa = 4.17
Molecular weight: 39.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3850 |
220 |
2175 |
770.0 |
88.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.571 ± 1.113 | 1.403 ± 0.294 |
5.792 ± 0.471 | 7.74 ± 0.396 |
4.338 ± 0.318 | 5.403 ± 0.43 |
2.208 ± 0.221 | 7.558 ± 0.381 |
7.325 ± 1.074 | 9.325 ± 1.123 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.468 ± 0.111 | 5.558 ± 0.209 |
3.948 ± 0.288 | 3.299 ± 0.64 |
5.377 ± 0.414 | 7.325 ± 0.922 |
5.844 ± 0.139 | 5.169 ± 0.553 |
1.506 ± 0.323 | 3.844 ± 0.288 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
