
Sanxia tombus-like virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 9.09
Get precalculated fractions of proteins
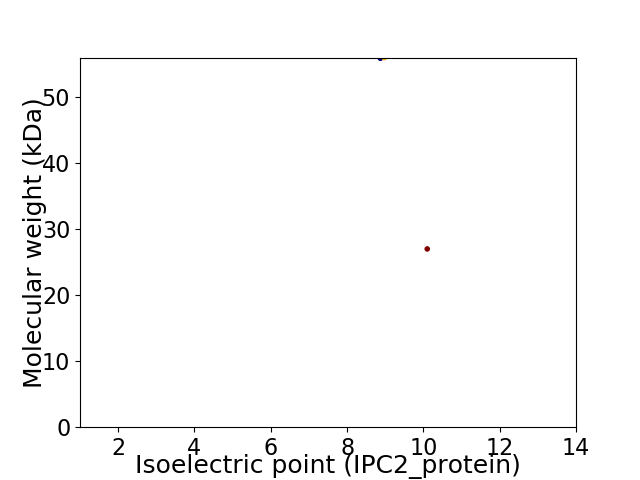
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGN8|A0A1L3KGN8_9VIRU Uncharacterized protein OS=Sanxia tombus-like virus 6 OX=1923390 PE=4 SV=1
MM1 pKa = 7.43SSAKK5 pKa = 9.95VVYY8 pKa = 10.01KK9 pKa = 10.4PSGHH13 pKa = 6.37CNEE16 pKa = 4.28SRR18 pKa = 11.84TAFTVIAALPSYY30 pKa = 10.11RR31 pKa = 11.84VAVHH35 pKa = 6.58KK36 pKa = 11.05NCICNEE42 pKa = 3.82LVSLHH47 pKa = 5.81NRR49 pKa = 11.84HH50 pKa = 6.35LRR52 pKa = 11.84QQAPGNQPYY61 pKa = 8.14TAAALSMWAPVYY73 pKa = 10.06DD74 pKa = 3.88VPRR77 pKa = 11.84ANIWDD82 pKa = 3.61IVKK85 pKa = 10.26GYY87 pKa = 10.78SGGKK91 pKa = 7.94RR92 pKa = 11.84KK93 pKa = 10.01AYY95 pKa = 10.22ARR97 pKa = 11.84AAINLQQPFQRR108 pKa = 11.84HH109 pKa = 4.09WAYY112 pKa = 10.72VSAFIKK118 pKa = 9.97PDD120 pKa = 4.11KK121 pKa = 10.89IPVDD125 pKa = 4.71EE126 pKa = 5.07IADD129 pKa = 3.73KK130 pKa = 10.97APRR133 pKa = 11.84LIQYY137 pKa = 9.77RR138 pKa = 11.84SKK140 pKa = 10.97EE141 pKa = 4.13YY142 pKa = 10.94NLLLATYY149 pKa = 8.51LKK151 pKa = 10.23PLEE154 pKa = 4.67DD155 pKa = 3.77AMFDD159 pKa = 3.54EE160 pKa = 5.02TDD162 pKa = 3.36PTGTLIFAKK171 pKa = 10.01KK172 pKa = 10.15RR173 pKa = 11.84NPQQRR178 pKa = 11.84AADD181 pKa = 4.36LLAKK185 pKa = 9.48MAAVEE190 pKa = 4.13DD191 pKa = 4.19ALVICADD198 pKa = 3.28HH199 pKa = 6.89SKK201 pKa = 10.6FDD203 pKa = 3.5SSVRR207 pKa = 11.84KK208 pKa = 8.04QHH210 pKa = 5.87LRR212 pKa = 11.84HH213 pKa = 6.21IEE215 pKa = 3.62AEE217 pKa = 4.11YY218 pKa = 10.18RR219 pKa = 11.84RR220 pKa = 11.84SYY222 pKa = 10.32PRR224 pKa = 11.84SKK226 pKa = 10.56LLKK229 pKa = 8.4RR230 pKa = 11.84LLRR233 pKa = 11.84LRR235 pKa = 11.84HH236 pKa = 5.02VNKK239 pKa = 10.06CRR241 pKa = 11.84TRR243 pKa = 11.84NGIKK247 pKa = 9.74YY248 pKa = 9.39QSKK251 pKa = 8.17EE252 pKa = 4.26TVQSGDD258 pKa = 3.17YY259 pKa = 9.43DD260 pKa = 3.54TGGRR264 pKa = 11.84NSRR267 pKa = 11.84LNAKK271 pKa = 9.61VLTSYY276 pKa = 10.9CILAGATTYY285 pKa = 11.42GLYY288 pKa = 9.72IDD290 pKa = 5.54GDD292 pKa = 4.13DD293 pKa = 4.2SVVIMPSALYY303 pKa = 10.18NEE305 pKa = 4.93KK306 pKa = 10.59AFKK309 pKa = 10.24DD310 pKa = 3.62HH311 pKa = 6.86CSKK314 pKa = 11.13LGFQTKK320 pKa = 9.26CQAYY324 pKa = 9.77RR325 pKa = 11.84DD326 pKa = 3.68LAEE329 pKa = 5.28ADD331 pKa = 3.83FCQAHH336 pKa = 7.56CIRR339 pKa = 11.84SDD341 pKa = 3.73PPTMMRR347 pKa = 11.84DD348 pKa = 3.71PLRR351 pKa = 11.84AAAHH355 pKa = 5.7FNISVRR361 pKa = 11.84NYY363 pKa = 10.42NGPAWPRR370 pKa = 11.84LVEE373 pKa = 4.2GKK375 pKa = 10.33LMCEE379 pKa = 3.78EE380 pKa = 4.06AAGRR384 pKa = 11.84GCPMMGKK391 pKa = 9.73LARR394 pKa = 11.84RR395 pKa = 11.84LRR397 pKa = 11.84TGIKK401 pKa = 10.04PIFDD405 pKa = 4.22PEE407 pKa = 4.41DD408 pKa = 3.46LEE410 pKa = 4.12KK411 pKa = 10.6WKK413 pKa = 10.57LVKK416 pKa = 10.36DD417 pKa = 4.25LPQATITMQARR428 pKa = 11.84LDD430 pKa = 3.95VYY432 pKa = 7.58TAWGFSVQDD441 pKa = 3.09QHH443 pKa = 7.71NFEE446 pKa = 4.45SMPRR450 pKa = 11.84HH451 pKa = 5.4CVVPEE456 pKa = 3.31IRR458 pKa = 11.84SRR460 pKa = 11.84RR461 pKa = 11.84NYY463 pKa = 10.04DD464 pKa = 3.11AEE466 pKa = 3.98SLHH469 pKa = 6.76RR470 pKa = 11.84VFEE473 pKa = 4.32AWSAMGATRR482 pKa = 11.84SEE484 pKa = 4.34SSWYY488 pKa = 10.39RR489 pKa = 11.84GTT491 pKa = 3.74
MM1 pKa = 7.43SSAKK5 pKa = 9.95VVYY8 pKa = 10.01KK9 pKa = 10.4PSGHH13 pKa = 6.37CNEE16 pKa = 4.28SRR18 pKa = 11.84TAFTVIAALPSYY30 pKa = 10.11RR31 pKa = 11.84VAVHH35 pKa = 6.58KK36 pKa = 11.05NCICNEE42 pKa = 3.82LVSLHH47 pKa = 5.81NRR49 pKa = 11.84HH50 pKa = 6.35LRR52 pKa = 11.84QQAPGNQPYY61 pKa = 8.14TAAALSMWAPVYY73 pKa = 10.06DD74 pKa = 3.88VPRR77 pKa = 11.84ANIWDD82 pKa = 3.61IVKK85 pKa = 10.26GYY87 pKa = 10.78SGGKK91 pKa = 7.94RR92 pKa = 11.84KK93 pKa = 10.01AYY95 pKa = 10.22ARR97 pKa = 11.84AAINLQQPFQRR108 pKa = 11.84HH109 pKa = 4.09WAYY112 pKa = 10.72VSAFIKK118 pKa = 9.97PDD120 pKa = 4.11KK121 pKa = 10.89IPVDD125 pKa = 4.71EE126 pKa = 5.07IADD129 pKa = 3.73KK130 pKa = 10.97APRR133 pKa = 11.84LIQYY137 pKa = 9.77RR138 pKa = 11.84SKK140 pKa = 10.97EE141 pKa = 4.13YY142 pKa = 10.94NLLLATYY149 pKa = 8.51LKK151 pKa = 10.23PLEE154 pKa = 4.67DD155 pKa = 3.77AMFDD159 pKa = 3.54EE160 pKa = 5.02TDD162 pKa = 3.36PTGTLIFAKK171 pKa = 10.01KK172 pKa = 10.15RR173 pKa = 11.84NPQQRR178 pKa = 11.84AADD181 pKa = 4.36LLAKK185 pKa = 9.48MAAVEE190 pKa = 4.13DD191 pKa = 4.19ALVICADD198 pKa = 3.28HH199 pKa = 6.89SKK201 pKa = 10.6FDD203 pKa = 3.5SSVRR207 pKa = 11.84KK208 pKa = 8.04QHH210 pKa = 5.87LRR212 pKa = 11.84HH213 pKa = 6.21IEE215 pKa = 3.62AEE217 pKa = 4.11YY218 pKa = 10.18RR219 pKa = 11.84RR220 pKa = 11.84SYY222 pKa = 10.32PRR224 pKa = 11.84SKK226 pKa = 10.56LLKK229 pKa = 8.4RR230 pKa = 11.84LLRR233 pKa = 11.84LRR235 pKa = 11.84HH236 pKa = 5.02VNKK239 pKa = 10.06CRR241 pKa = 11.84TRR243 pKa = 11.84NGIKK247 pKa = 9.74YY248 pKa = 9.39QSKK251 pKa = 8.17EE252 pKa = 4.26TVQSGDD258 pKa = 3.17YY259 pKa = 9.43DD260 pKa = 3.54TGGRR264 pKa = 11.84NSRR267 pKa = 11.84LNAKK271 pKa = 9.61VLTSYY276 pKa = 10.9CILAGATTYY285 pKa = 11.42GLYY288 pKa = 9.72IDD290 pKa = 5.54GDD292 pKa = 4.13DD293 pKa = 4.2SVVIMPSALYY303 pKa = 10.18NEE305 pKa = 4.93KK306 pKa = 10.59AFKK309 pKa = 10.24DD310 pKa = 3.62HH311 pKa = 6.86CSKK314 pKa = 11.13LGFQTKK320 pKa = 9.26CQAYY324 pKa = 9.77RR325 pKa = 11.84DD326 pKa = 3.68LAEE329 pKa = 5.28ADD331 pKa = 3.83FCQAHH336 pKa = 7.56CIRR339 pKa = 11.84SDD341 pKa = 3.73PPTMMRR347 pKa = 11.84DD348 pKa = 3.71PLRR351 pKa = 11.84AAAHH355 pKa = 5.7FNISVRR361 pKa = 11.84NYY363 pKa = 10.42NGPAWPRR370 pKa = 11.84LVEE373 pKa = 4.2GKK375 pKa = 10.33LMCEE379 pKa = 3.78EE380 pKa = 4.06AAGRR384 pKa = 11.84GCPMMGKK391 pKa = 9.73LARR394 pKa = 11.84RR395 pKa = 11.84LRR397 pKa = 11.84TGIKK401 pKa = 10.04PIFDD405 pKa = 4.22PEE407 pKa = 4.41DD408 pKa = 3.46LEE410 pKa = 4.12KK411 pKa = 10.6WKK413 pKa = 10.57LVKK416 pKa = 10.36DD417 pKa = 4.25LPQATITMQARR428 pKa = 11.84LDD430 pKa = 3.95VYY432 pKa = 7.58TAWGFSVQDD441 pKa = 3.09QHH443 pKa = 7.71NFEE446 pKa = 4.45SMPRR450 pKa = 11.84HH451 pKa = 5.4CVVPEE456 pKa = 3.31IRR458 pKa = 11.84SRR460 pKa = 11.84RR461 pKa = 11.84NYY463 pKa = 10.04DD464 pKa = 3.11AEE466 pKa = 3.98SLHH469 pKa = 6.76RR470 pKa = 11.84VFEE473 pKa = 4.32AWSAMGATRR482 pKa = 11.84SEE484 pKa = 4.34SSWYY488 pKa = 10.39RR489 pKa = 11.84GTT491 pKa = 3.74
Molecular weight: 55.85 kDa
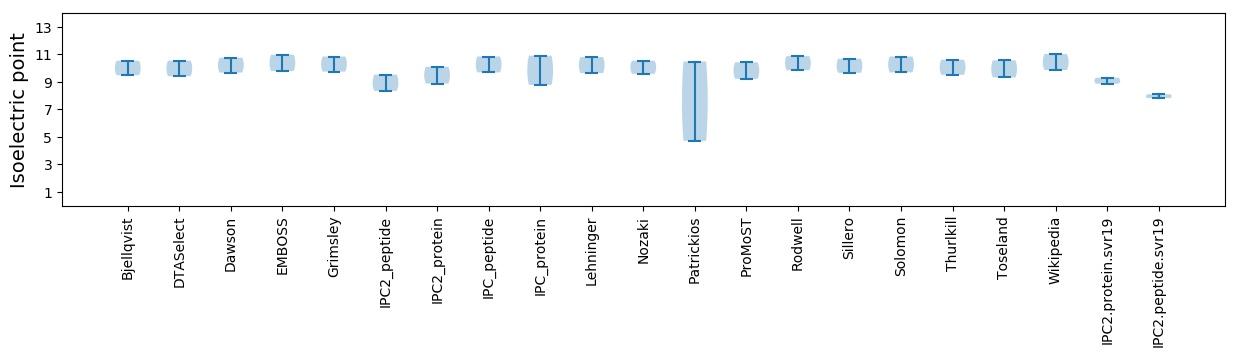
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGN8|A0A1L3KGN8_9VIRU Uncharacterized protein OS=Sanxia tombus-like virus 6 OX=1923390 PE=4 SV=1
MM1 pKa = 7.46TLNPYY6 pKa = 8.86TGYY9 pKa = 10.5SRR11 pKa = 11.84RR12 pKa = 11.84GVPWALLGARR22 pKa = 11.84AAGTVARR29 pKa = 11.84NAWQWYY35 pKa = 5.9NQPPAAPMYY44 pKa = 9.52GPPIPPPRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84QAPVYY60 pKa = 9.75APQPGPSQAAPRR72 pKa = 11.84GRR74 pKa = 11.84GRR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84PRR81 pKa = 11.84GTGAPSRR88 pKa = 11.84PLATTKK94 pKa = 10.46EE95 pKa = 4.1GSNLVVRR102 pKa = 11.84DD103 pKa = 3.77TEE105 pKa = 4.57IISLGAKK112 pKa = 7.73NTLFTMQFKK121 pKa = 10.36PSEE124 pKa = 4.3TTLPRR129 pKa = 11.84LDD131 pKa = 3.91KK132 pKa = 11.27EE133 pKa = 4.23GGAFNRR139 pKa = 11.84YY140 pKa = 9.19RR141 pKa = 11.84IQYY144 pKa = 9.65VNIAYY149 pKa = 9.71ISTSSAATAGSVSYY163 pKa = 10.78GVLQGPTNANIATLNAIVKK182 pKa = 8.35LRR184 pKa = 11.84PMEE187 pKa = 4.25SHH189 pKa = 6.67AVWKK193 pKa = 10.7NSSLSLGSDD202 pKa = 2.66IMSAKK207 pKa = 10.06YY208 pKa = 10.51LSTAATGEE216 pKa = 4.11DD217 pKa = 3.06AVAFTVYY224 pKa = 10.2AWNTADD230 pKa = 4.14SPGYY234 pKa = 9.82LQFSYY239 pKa = 10.07RR240 pKa = 11.84VEE242 pKa = 4.01FAFPKK247 pKa = 10.03PP248 pKa = 3.48
MM1 pKa = 7.46TLNPYY6 pKa = 8.86TGYY9 pKa = 10.5SRR11 pKa = 11.84RR12 pKa = 11.84GVPWALLGARR22 pKa = 11.84AAGTVARR29 pKa = 11.84NAWQWYY35 pKa = 5.9NQPPAAPMYY44 pKa = 9.52GPPIPPPRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84QAPVYY60 pKa = 9.75APQPGPSQAAPRR72 pKa = 11.84GRR74 pKa = 11.84GRR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84PRR81 pKa = 11.84GTGAPSRR88 pKa = 11.84PLATTKK94 pKa = 10.46EE95 pKa = 4.1GSNLVVRR102 pKa = 11.84DD103 pKa = 3.77TEE105 pKa = 4.57IISLGAKK112 pKa = 7.73NTLFTMQFKK121 pKa = 10.36PSEE124 pKa = 4.3TTLPRR129 pKa = 11.84LDD131 pKa = 3.91KK132 pKa = 11.27EE133 pKa = 4.23GGAFNRR139 pKa = 11.84YY140 pKa = 9.19RR141 pKa = 11.84IQYY144 pKa = 9.65VNIAYY149 pKa = 9.71ISTSSAATAGSVSYY163 pKa = 10.78GVLQGPTNANIATLNAIVKK182 pKa = 8.35LRR184 pKa = 11.84PMEE187 pKa = 4.25SHH189 pKa = 6.67AVWKK193 pKa = 10.7NSSLSLGSDD202 pKa = 2.66IMSAKK207 pKa = 10.06YY208 pKa = 10.51LSTAATGEE216 pKa = 4.11DD217 pKa = 3.06AVAFTVYY224 pKa = 10.2AWNTADD230 pKa = 4.14SPGYY234 pKa = 9.82LQFSYY239 pKa = 10.07RR240 pKa = 11.84VEE242 pKa = 4.01FAFPKK247 pKa = 10.03PP248 pKa = 3.48
Molecular weight: 26.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
739 |
248 |
491 |
369.5 |
41.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.502 ± 0.73 | 1.759 ± 0.917 |
4.465 ± 1.276 | 3.924 ± 0.574 |
2.842 ± 0.01 | 5.819 ± 1.17 |
2.165 ± 0.918 | 4.195 ± 0.295 |
5.413 ± 1.139 | 7.442 ± 0.516 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.436 ± 0.219 | 4.195 ± 0.335 |
6.901 ± 1.657 | 3.789 ± 0.083 |
8.66 ± 0.11 | 7.037 ± 0.536 |
5.413 ± 1.172 | 5.413 ± 0.089 |
1.759 ± 0.134 | 4.871 ± 0.193 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
