
Pseudolysobacter antarcticus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Pseudolysobacter
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
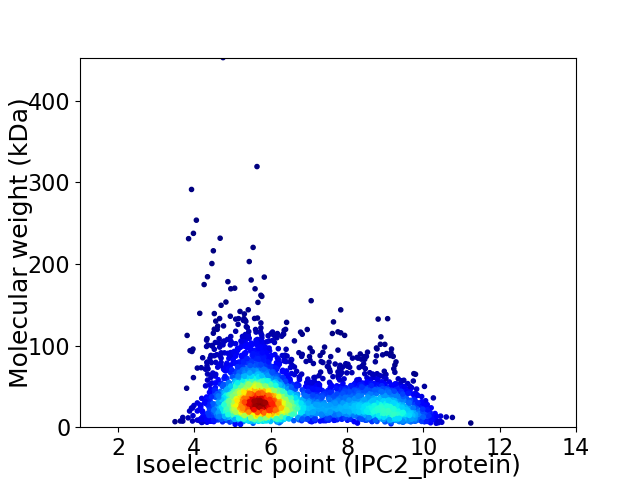
Virtual 2D-PAGE plot for 3894 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
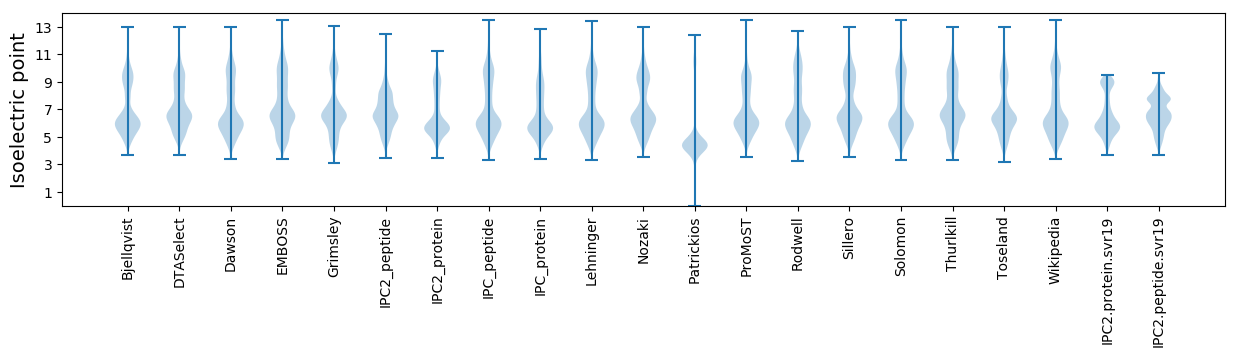
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A411HLQ0|A0A411HLQ0_9GAMM Ribosome maturation factor RimP OS=Pseudolysobacter antarcticus OX=2511995 GN=rimP PE=3 SV=1
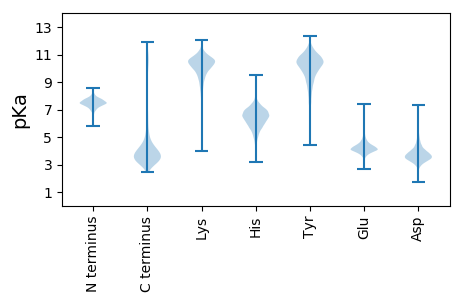
MM1 pKa = 7.44NIIHH5 pKa = 7.03FLRR8 pKa = 11.84GVPRR12 pKa = 11.84RR13 pKa = 11.84FAAFVFGICATLPAFADD30 pKa = 3.68SSTLTISAASVHH42 pKa = 6.28VDD44 pKa = 3.31NAPTTLYY51 pKa = 10.77FPITRR56 pKa = 11.84SGDD59 pKa = 3.01ISYY62 pKa = 8.12DD63 pKa = 3.28TVLNYY68 pKa = 8.76HH69 pKa = 5.82TVDD72 pKa = 3.58GSAMAGIDD80 pKa = 3.43YY81 pKa = 7.64TANTGSVSLLSGSSSASIPVTITPSAPTSASNSFKK116 pKa = 10.44MLLDD120 pKa = 3.34STINLIAPGFAAHH133 pKa = 6.53VEE135 pKa = 4.27TSLGFDD141 pKa = 3.74VYY143 pKa = 11.41ALSTADD149 pKa = 4.09LNGDD153 pKa = 4.38GKK155 pKa = 10.04PDD157 pKa = 3.77LAIVSYY163 pKa = 11.07GSDD166 pKa = 2.75KK167 pKa = 11.23VSFLLNTTPAGAATPSFGAVQSINAGCAPQSIITADD203 pKa = 3.18INGDD207 pKa = 3.63GKK209 pKa = 10.12PDD211 pKa = 3.41VVFACPFDD219 pKa = 3.96NKK221 pKa = 9.24VTLLMNTTATGASTTSFATQVFTVAGPSSLKK252 pKa = 10.24ATDD255 pKa = 4.19LNGDD259 pKa = 3.66NKK261 pKa = 10.85LDD263 pKa = 5.51LIVTDD268 pKa = 3.65TNNYY272 pKa = 8.19QVVVMLNSTTVDD284 pKa = 3.04ASSASFFGTQAFATGMVPDD303 pKa = 6.28AITSTDD309 pKa = 2.98INGDD313 pKa = 3.66GKK315 pKa = 10.17PDD317 pKa = 3.42LVVSNRR323 pKa = 11.84LSNSLSVLLNTTANGAATASFAAQQVFATGTNPVWVSAIDD363 pKa = 3.4INGDD367 pKa = 3.65GKK369 pKa = 10.18PDD371 pKa = 3.56LAVANYY377 pKa = 9.69GSSTFSILLNTTTTGAAIPSFAPQQIFPTGSHH409 pKa = 6.05PTSIVSADD417 pKa = 3.45VNSDD421 pKa = 2.97GKK423 pKa = 9.47TDD425 pKa = 4.13LLVTNSFDD433 pKa = 3.31NTLSVFSNYY442 pKa = 7.37TAIGSSTAGFTINLYY457 pKa = 9.16ATGFSPQSGFAIDD470 pKa = 4.53LNADD474 pKa = 3.08GRR476 pKa = 11.84LDD478 pKa = 4.09LLTVNASGGDD488 pKa = 3.45VSVLMNTTVPPALSFASMQNLGTGGYY514 pKa = 8.09TNGVTSADD522 pKa = 3.27INGDD526 pKa = 3.35GKK528 pKa = 11.29LDD530 pKa = 4.64LIEE533 pKa = 4.48VDD535 pKa = 4.05RR536 pKa = 11.84SDD538 pKa = 3.31NTVSVFINTTPAGSATTSYY557 pKa = 11.22APRR560 pKa = 11.84RR561 pKa = 11.84TFATSSLPWAVAVADD576 pKa = 4.5LNGDD580 pKa = 4.29GKK582 pKa = 10.18PDD584 pKa = 3.04IAVANFFGSSVSVLTNQTATGATTASFSAQATFNTGSSPISIAITDD630 pKa = 3.52INGDD634 pKa = 3.56GKK636 pKa = 11.29LDD638 pKa = 3.6ILVADD643 pKa = 5.01AGANDD648 pKa = 3.61VAVLRR653 pKa = 11.84NTTTTGAATPSFAAYY668 pKa = 10.4LSLPAGGAPSAVASADD684 pKa = 3.55LNGDD688 pKa = 3.59GKK690 pKa = 11.36SDD692 pKa = 3.01IVVTNKK698 pKa = 10.44DD699 pKa = 3.64DD700 pKa = 3.77GTVSVFQNTTTVATTPTFAAQQILSVGSQPYY731 pKa = 9.75SVAIADD737 pKa = 3.84FNGDD741 pKa = 3.21GKK743 pKa = 11.0RR744 pKa = 11.84DD745 pKa = 3.65VVVANYY751 pKa = 10.23ASSTVSILLNTTTAPGATVSLAAQQTFDD779 pKa = 3.71GGNPYY784 pKa = 10.92AVTTADD790 pKa = 3.24INGDD794 pKa = 3.68GKK796 pKa = 10.1PDD798 pKa = 3.66VIVAVPGNSTAAVLLNTSASGTLNFASPMYY828 pKa = 10.33LSIGGSPTAIATTDD842 pKa = 3.33ANSDD846 pKa = 3.4GRR848 pKa = 11.84VDD850 pKa = 3.28IVSTNGGSNSLSILLNSQILTNLAGSPATGTIVHH884 pKa = 6.79DD885 pKa = 5.21LIVTPSAGEE894 pKa = 3.87NGSISQSSPQTVAYY908 pKa = 7.23GTSTSFTVTPLPNYY922 pKa = 8.64VATVVGTCGGTLLGTTYY939 pKa = 7.88TTAAITANCTVIASFAAVTHH959 pKa = 6.07IVMPSADD966 pKa = 3.4AHH968 pKa = 6.26GSISPNMAQNIDD980 pKa = 3.75HH981 pKa = 7.17GSTTMFTIMPDD992 pKa = 3.05TGYY995 pKa = 10.3SASVSGTCGGTLVDD1009 pKa = 4.24ATYY1012 pKa = 7.29TTNAITTDD1020 pKa = 3.69CSVVASFTLKK1030 pKa = 10.53SYY1032 pKa = 11.02LVTPSVGDD1040 pKa = 3.63YY1041 pKa = 11.4GSISPDD1047 pKa = 2.98TPQNIDD1053 pKa = 3.29YY1054 pKa = 10.21GATTSFMVTPNAGFIVIVGGTCGGTLVDD1082 pKa = 3.96STYY1085 pKa = 8.1TTNTITANCTVAVTFSDD1102 pKa = 4.89RR1103 pKa = 11.84IFADD1107 pKa = 4.05GFEE1110 pKa = 4.28QPP1112 pKa = 4.2
MM1 pKa = 7.44NIIHH5 pKa = 7.03FLRR8 pKa = 11.84GVPRR12 pKa = 11.84RR13 pKa = 11.84FAAFVFGICATLPAFADD30 pKa = 3.68SSTLTISAASVHH42 pKa = 6.28VDD44 pKa = 3.31NAPTTLYY51 pKa = 10.77FPITRR56 pKa = 11.84SGDD59 pKa = 3.01ISYY62 pKa = 8.12DD63 pKa = 3.28TVLNYY68 pKa = 8.76HH69 pKa = 5.82TVDD72 pKa = 3.58GSAMAGIDD80 pKa = 3.43YY81 pKa = 7.64TANTGSVSLLSGSSSASIPVTITPSAPTSASNSFKK116 pKa = 10.44MLLDD120 pKa = 3.34STINLIAPGFAAHH133 pKa = 6.53VEE135 pKa = 4.27TSLGFDD141 pKa = 3.74VYY143 pKa = 11.41ALSTADD149 pKa = 4.09LNGDD153 pKa = 4.38GKK155 pKa = 10.04PDD157 pKa = 3.77LAIVSYY163 pKa = 11.07GSDD166 pKa = 2.75KK167 pKa = 11.23VSFLLNTTPAGAATPSFGAVQSINAGCAPQSIITADD203 pKa = 3.18INGDD207 pKa = 3.63GKK209 pKa = 10.12PDD211 pKa = 3.41VVFACPFDD219 pKa = 3.96NKK221 pKa = 9.24VTLLMNTTATGASTTSFATQVFTVAGPSSLKK252 pKa = 10.24ATDD255 pKa = 4.19LNGDD259 pKa = 3.66NKK261 pKa = 10.85LDD263 pKa = 5.51LIVTDD268 pKa = 3.65TNNYY272 pKa = 8.19QVVVMLNSTTVDD284 pKa = 3.04ASSASFFGTQAFATGMVPDD303 pKa = 6.28AITSTDD309 pKa = 2.98INGDD313 pKa = 3.66GKK315 pKa = 10.17PDD317 pKa = 3.42LVVSNRR323 pKa = 11.84LSNSLSVLLNTTANGAATASFAAQQVFATGTNPVWVSAIDD363 pKa = 3.4INGDD367 pKa = 3.65GKK369 pKa = 10.18PDD371 pKa = 3.56LAVANYY377 pKa = 9.69GSSTFSILLNTTTTGAAIPSFAPQQIFPTGSHH409 pKa = 6.05PTSIVSADD417 pKa = 3.45VNSDD421 pKa = 2.97GKK423 pKa = 9.47TDD425 pKa = 4.13LLVTNSFDD433 pKa = 3.31NTLSVFSNYY442 pKa = 7.37TAIGSSTAGFTINLYY457 pKa = 9.16ATGFSPQSGFAIDD470 pKa = 4.53LNADD474 pKa = 3.08GRR476 pKa = 11.84LDD478 pKa = 4.09LLTVNASGGDD488 pKa = 3.45VSVLMNTTVPPALSFASMQNLGTGGYY514 pKa = 8.09TNGVTSADD522 pKa = 3.27INGDD526 pKa = 3.35GKK528 pKa = 11.29LDD530 pKa = 4.64LIEE533 pKa = 4.48VDD535 pKa = 4.05RR536 pKa = 11.84SDD538 pKa = 3.31NTVSVFINTTPAGSATTSYY557 pKa = 11.22APRR560 pKa = 11.84RR561 pKa = 11.84TFATSSLPWAVAVADD576 pKa = 4.5LNGDD580 pKa = 4.29GKK582 pKa = 10.18PDD584 pKa = 3.04IAVANFFGSSVSVLTNQTATGATTASFSAQATFNTGSSPISIAITDD630 pKa = 3.52INGDD634 pKa = 3.56GKK636 pKa = 11.29LDD638 pKa = 3.6ILVADD643 pKa = 5.01AGANDD648 pKa = 3.61VAVLRR653 pKa = 11.84NTTTTGAATPSFAAYY668 pKa = 10.4LSLPAGGAPSAVASADD684 pKa = 3.55LNGDD688 pKa = 3.59GKK690 pKa = 11.36SDD692 pKa = 3.01IVVTNKK698 pKa = 10.44DD699 pKa = 3.64DD700 pKa = 3.77GTVSVFQNTTTVATTPTFAAQQILSVGSQPYY731 pKa = 9.75SVAIADD737 pKa = 3.84FNGDD741 pKa = 3.21GKK743 pKa = 11.0RR744 pKa = 11.84DD745 pKa = 3.65VVVANYY751 pKa = 10.23ASSTVSILLNTTTAPGATVSLAAQQTFDD779 pKa = 3.71GGNPYY784 pKa = 10.92AVTTADD790 pKa = 3.24INGDD794 pKa = 3.68GKK796 pKa = 10.1PDD798 pKa = 3.66VIVAVPGNSTAAVLLNTSASGTLNFASPMYY828 pKa = 10.33LSIGGSPTAIATTDD842 pKa = 3.33ANSDD846 pKa = 3.4GRR848 pKa = 11.84VDD850 pKa = 3.28IVSTNGGSNSLSILLNSQILTNLAGSPATGTIVHH884 pKa = 6.79DD885 pKa = 5.21LIVTPSAGEE894 pKa = 3.87NGSISQSSPQTVAYY908 pKa = 7.23GTSTSFTVTPLPNYY922 pKa = 8.64VATVVGTCGGTLLGTTYY939 pKa = 7.88TTAAITANCTVIASFAAVTHH959 pKa = 6.07IVMPSADD966 pKa = 3.4AHH968 pKa = 6.26GSISPNMAQNIDD980 pKa = 3.75HH981 pKa = 7.17GSTTMFTIMPDD992 pKa = 3.05TGYY995 pKa = 10.3SASVSGTCGGTLVDD1009 pKa = 4.24ATYY1012 pKa = 7.29TTNAITTDD1020 pKa = 3.69CSVVASFTLKK1030 pKa = 10.53SYY1032 pKa = 11.02LVTPSVGDD1040 pKa = 3.63YY1041 pKa = 11.4GSISPDD1047 pKa = 2.98TPQNIDD1053 pKa = 3.29YY1054 pKa = 10.21GATTSFMVTPNAGFIVIVGGTCGGTLVDD1082 pKa = 3.96STYY1085 pKa = 8.1TTNTITANCTVAVTFSDD1102 pKa = 4.89RR1103 pKa = 11.84IFADD1107 pKa = 4.05GFEE1110 pKa = 4.28QPP1112 pKa = 4.2
Molecular weight: 112.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A411HPS7|A0A411HPS7_9GAMM Patatin-like phospholipase family protein OS=Pseudolysobacter antarcticus OX=2511995 GN=ELE36_20075 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.05GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 9.02TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.61GRR39 pKa = 11.84KK40 pKa = 9.07RR41 pKa = 11.84LTPP44 pKa = 3.95
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.05GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 9.02TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.61GRR39 pKa = 11.84KK40 pKa = 9.07RR41 pKa = 11.84LTPP44 pKa = 3.95
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1372272 |
30 |
4690 |
352.4 |
38.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.178 ± 0.051 | 0.972 ± 0.015 |
5.533 ± 0.028 | 4.716 ± 0.051 |
3.738 ± 0.024 | 7.961 ± 0.052 |
2.365 ± 0.025 | 5.282 ± 0.027 |
3.19 ± 0.036 | 10.676 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.05 ± 0.022 | 3.459 ± 0.04 |
4.876 ± 0.024 | 3.788 ± 0.026 |
6.305 ± 0.056 | 6.316 ± 0.045 |
5.748 ± 0.076 | 6.967 ± 0.031 |
1.365 ± 0.018 | 2.515 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
