
Phaeobacter gallaeciensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae;
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
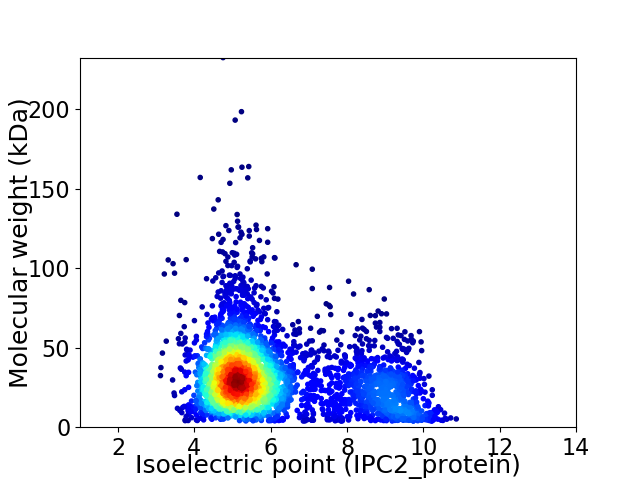
Virtual 2D-PAGE plot for 3897 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B0ZND8|A0A1B0ZND8_9RHOB Bcr/CflA family efflux transporter OS=Phaeobacter gallaeciensis OX=60890 GN=bcr PE=3 SV=1
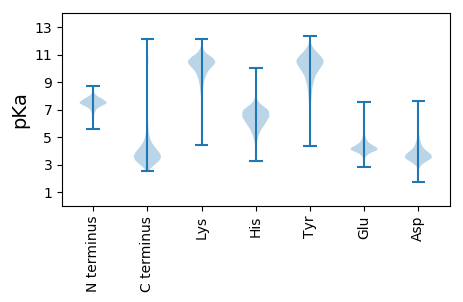
MM1 pKa = 7.28NKK3 pKa = 8.0ITGAFIAALALSACGNGNPFTEE25 pKa = 4.52DD26 pKa = 2.98TTDD29 pKa = 3.15NGGDD33 pKa = 3.37TGTDD37 pKa = 2.91TGTDD41 pKa = 3.16TGIDD45 pKa = 3.63GDD47 pKa = 4.28GRR49 pKa = 11.84PPGTPSPTPDD59 pKa = 2.76TSIFRR64 pKa = 11.84RR65 pKa = 11.84EE66 pKa = 4.04GTSTEE71 pKa = 3.83DD72 pKa = 3.18AYY74 pKa = 10.18IGNGYY79 pKa = 8.13ATEE82 pKa = 3.93ISYY85 pKa = 11.15DD86 pKa = 3.77SNNDD90 pKa = 3.06TFSVDD95 pKa = 3.11NLAFDD100 pKa = 4.71GDD102 pKa = 3.69GTYY105 pKa = 11.19ARR107 pKa = 11.84GTAVSSLGPFAVYY120 pKa = 9.77EE121 pKa = 4.09ADD123 pKa = 3.42AQYY126 pKa = 11.66SDD128 pKa = 3.31ITDD131 pKa = 3.61NEE133 pKa = 4.75PINQFTHH140 pKa = 6.18RR141 pKa = 11.84AIYY144 pKa = 9.89GVSTSGNSKK153 pKa = 9.89FAIVRR158 pKa = 11.84TGAYY162 pKa = 9.9ADD164 pKa = 3.58YY165 pKa = 10.8GFGGFVYY172 pKa = 10.54QRR174 pKa = 11.84DD175 pKa = 3.71NSVTLPSSGQAQYY188 pKa = 11.12NGSLAGLRR196 pKa = 11.84DD197 pKa = 3.75YY198 pKa = 11.59NGAAGLEE205 pKa = 4.4YY206 pKa = 8.59TTADD210 pKa = 3.33VQIAIDD216 pKa = 4.21FDD218 pKa = 4.34DD219 pKa = 5.04FNPATGGRR227 pKa = 11.84GDD229 pKa = 3.67AVRR232 pKa = 11.84GSLTNRR238 pKa = 11.84TIYY241 pKa = 9.46DD242 pKa = 3.48TAGNDD247 pKa = 3.0ITNDD251 pKa = 3.17VINRR255 pKa = 11.84INVDD259 pKa = 3.24NNLTLSSIPVAVFDD273 pKa = 4.62VGPNNLDD280 pKa = 3.65DD281 pKa = 4.16NGEE284 pKa = 4.33IIGTLQSTYY293 pKa = 10.87VDD295 pKa = 3.6ANGTAQTYY303 pKa = 7.3EE304 pKa = 3.9TGNYY308 pKa = 8.01YY309 pKa = 10.9AVVSGDD315 pKa = 3.32NAEE318 pKa = 4.49EE319 pKa = 3.94IVGVVVLEE327 pKa = 4.28NSLDD331 pKa = 3.58PAANSVRR338 pKa = 11.84EE339 pKa = 3.96TGGFIVYY346 pKa = 10.64NNN348 pKa = 3.49
MM1 pKa = 7.28NKK3 pKa = 8.0ITGAFIAALALSACGNGNPFTEE25 pKa = 4.52DD26 pKa = 2.98TTDD29 pKa = 3.15NGGDD33 pKa = 3.37TGTDD37 pKa = 2.91TGTDD41 pKa = 3.16TGIDD45 pKa = 3.63GDD47 pKa = 4.28GRR49 pKa = 11.84PPGTPSPTPDD59 pKa = 2.76TSIFRR64 pKa = 11.84RR65 pKa = 11.84EE66 pKa = 4.04GTSTEE71 pKa = 3.83DD72 pKa = 3.18AYY74 pKa = 10.18IGNGYY79 pKa = 8.13ATEE82 pKa = 3.93ISYY85 pKa = 11.15DD86 pKa = 3.77SNNDD90 pKa = 3.06TFSVDD95 pKa = 3.11NLAFDD100 pKa = 4.71GDD102 pKa = 3.69GTYY105 pKa = 11.19ARR107 pKa = 11.84GTAVSSLGPFAVYY120 pKa = 9.77EE121 pKa = 4.09ADD123 pKa = 3.42AQYY126 pKa = 11.66SDD128 pKa = 3.31ITDD131 pKa = 3.61NEE133 pKa = 4.75PINQFTHH140 pKa = 6.18RR141 pKa = 11.84AIYY144 pKa = 9.89GVSTSGNSKK153 pKa = 9.89FAIVRR158 pKa = 11.84TGAYY162 pKa = 9.9ADD164 pKa = 3.58YY165 pKa = 10.8GFGGFVYY172 pKa = 10.54QRR174 pKa = 11.84DD175 pKa = 3.71NSVTLPSSGQAQYY188 pKa = 11.12NGSLAGLRR196 pKa = 11.84DD197 pKa = 3.75YY198 pKa = 11.59NGAAGLEE205 pKa = 4.4YY206 pKa = 8.59TTADD210 pKa = 3.33VQIAIDD216 pKa = 4.21FDD218 pKa = 4.34DD219 pKa = 5.04FNPATGGRR227 pKa = 11.84GDD229 pKa = 3.67AVRR232 pKa = 11.84GSLTNRR238 pKa = 11.84TIYY241 pKa = 9.46DD242 pKa = 3.48TAGNDD247 pKa = 3.0ITNDD251 pKa = 3.17VINRR255 pKa = 11.84INVDD259 pKa = 3.24NNLTLSSIPVAVFDD273 pKa = 4.62VGPNNLDD280 pKa = 3.65DD281 pKa = 4.16NGEE284 pKa = 4.33IIGTLQSTYY293 pKa = 10.87VDD295 pKa = 3.6ANGTAQTYY303 pKa = 7.3EE304 pKa = 3.9TGNYY308 pKa = 8.01YY309 pKa = 10.9AVVSGDD315 pKa = 3.32NAEE318 pKa = 4.49EE319 pKa = 3.94IVGVVVLEE327 pKa = 4.28NSLDD331 pKa = 3.58PAANSVRR338 pKa = 11.84EE339 pKa = 3.96TGGFIVYY346 pKa = 10.64NNN348 pKa = 3.49
Molecular weight: 36.47 kDa
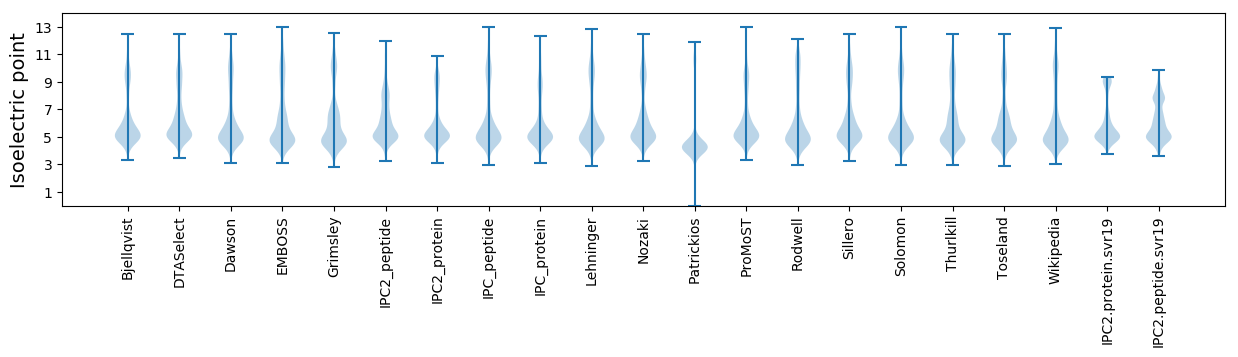
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B0ZP72|A0A1B0ZP72_9RHOB LysR family transcriptional regulator OS=Phaeobacter gallaeciensis OX=60890 GN=JL2886_00986 PE=3 SV=1
MM1 pKa = 7.34TEE3 pKa = 3.71QVVIVSPSGVARR15 pKa = 11.84LQRR18 pKa = 11.84EE19 pKa = 4.35AEE21 pKa = 4.13RR22 pKa = 11.84IAAIKK27 pKa = 10.5ARR29 pKa = 11.84GAVPRR34 pKa = 11.84QCGDD38 pKa = 4.81AIPEE42 pKa = 4.15APARR46 pKa = 11.84GAFQVFEE53 pKa = 4.34PMRR56 pKa = 11.84LCPVGADD63 pKa = 3.83DD64 pKa = 6.35YY65 pKa = 10.7EE66 pKa = 4.39AQPVGYY72 pKa = 9.25RR73 pKa = 11.84GRR75 pKa = 11.84SAIRR79 pKa = 11.84NADD82 pKa = 3.42VFDD85 pKa = 3.96VMAAAAARR93 pKa = 11.84KK94 pKa = 8.96KK95 pKa = 10.57KK96 pKa = 9.59PAPFTLGQIAMGRR109 pKa = 11.84HH110 pKa = 4.6YY111 pKa = 10.69RR112 pKa = 11.84DD113 pKa = 2.88LVEE116 pKa = 4.3RR117 pKa = 11.84HH118 pKa = 4.79ATAGVRR124 pKa = 11.84CSSVEE129 pKa = 3.74ALRR132 pKa = 11.84TGGSGQGGEE141 pKa = 4.81FIDD144 pKa = 4.16AVLRR148 pKa = 11.84DD149 pKa = 3.78RR150 pKa = 11.84DD151 pKa = 4.12EE152 pKa = 5.09IDD154 pKa = 3.02LLRR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84IGTGSAIEE167 pKa = 4.06VRR169 pKa = 11.84RR170 pKa = 11.84VRR172 pKa = 11.84PSKK175 pKa = 10.12RR176 pKa = 11.84GSRR179 pKa = 11.84VSITNRR185 pKa = 11.84RR186 pKa = 11.84LVDD189 pKa = 3.38MVCIEE194 pKa = 4.17DD195 pKa = 3.44RR196 pKa = 11.84SLRR199 pKa = 11.84EE200 pKa = 3.61VLEE203 pKa = 3.83AHH205 pKa = 6.36GWSVYY210 pKa = 10.88GEE212 pKa = 4.15TVKK215 pKa = 10.76AVRR218 pKa = 11.84AALASVLDD226 pKa = 4.22RR227 pKa = 11.84MAGPVRR233 pKa = 11.84TGRR236 pKa = 11.84IHH238 pKa = 6.27TLHH241 pKa = 6.87KK242 pKa = 10.63GAA244 pKa = 5.08
MM1 pKa = 7.34TEE3 pKa = 3.71QVVIVSPSGVARR15 pKa = 11.84LQRR18 pKa = 11.84EE19 pKa = 4.35AEE21 pKa = 4.13RR22 pKa = 11.84IAAIKK27 pKa = 10.5ARR29 pKa = 11.84GAVPRR34 pKa = 11.84QCGDD38 pKa = 4.81AIPEE42 pKa = 4.15APARR46 pKa = 11.84GAFQVFEE53 pKa = 4.34PMRR56 pKa = 11.84LCPVGADD63 pKa = 3.83DD64 pKa = 6.35YY65 pKa = 10.7EE66 pKa = 4.39AQPVGYY72 pKa = 9.25RR73 pKa = 11.84GRR75 pKa = 11.84SAIRR79 pKa = 11.84NADD82 pKa = 3.42VFDD85 pKa = 3.96VMAAAAARR93 pKa = 11.84KK94 pKa = 8.96KK95 pKa = 10.57KK96 pKa = 9.59PAPFTLGQIAMGRR109 pKa = 11.84HH110 pKa = 4.6YY111 pKa = 10.69RR112 pKa = 11.84DD113 pKa = 2.88LVEE116 pKa = 4.3RR117 pKa = 11.84HH118 pKa = 4.79ATAGVRR124 pKa = 11.84CSSVEE129 pKa = 3.74ALRR132 pKa = 11.84TGGSGQGGEE141 pKa = 4.81FIDD144 pKa = 4.16AVLRR148 pKa = 11.84DD149 pKa = 3.78RR150 pKa = 11.84DD151 pKa = 4.12EE152 pKa = 5.09IDD154 pKa = 3.02LLRR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84IGTGSAIEE167 pKa = 4.06VRR169 pKa = 11.84RR170 pKa = 11.84VRR172 pKa = 11.84PSKK175 pKa = 10.12RR176 pKa = 11.84GSRR179 pKa = 11.84VSITNRR185 pKa = 11.84RR186 pKa = 11.84LVDD189 pKa = 3.38MVCIEE194 pKa = 4.17DD195 pKa = 3.44RR196 pKa = 11.84SLRR199 pKa = 11.84EE200 pKa = 3.61VLEE203 pKa = 3.83AHH205 pKa = 6.36GWSVYY210 pKa = 10.88GEE212 pKa = 4.15TVKK215 pKa = 10.76AVRR218 pKa = 11.84AALASVLDD226 pKa = 4.22RR227 pKa = 11.84MAGPVRR233 pKa = 11.84TGRR236 pKa = 11.84IHH238 pKa = 6.27TLHH241 pKa = 6.87KK242 pKa = 10.63GAA244 pKa = 5.08
Molecular weight: 26.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1197673 |
37 |
2173 |
307.3 |
33.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.156 ± 0.048 | 0.915 ± 0.013 |
6.016 ± 0.037 | 6.147 ± 0.036 |
3.659 ± 0.023 | 8.705 ± 0.039 |
2.06 ± 0.022 | 5.096 ± 0.032 |
3.351 ± 0.032 | 10.123 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.891 ± 0.019 | 2.624 ± 0.024 |
5.012 ± 0.031 | 3.485 ± 0.024 |
6.394 ± 0.039 | 5.344 ± 0.029 |
5.386 ± 0.029 | 7.068 ± 0.035 |
1.346 ± 0.016 | 2.223 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
